Low-level TP53 mutational load antecedes clonal expansion in chronic lymphocytic leukaemia
The presence of still unknown genetic and epigenetic alterations and antigenic stimulation may lead to the selection and expansion of functionally inert mature B cells, with the generation of oligoclonal populations in chronic lymphocytic leukaemia (CLL) (Knittel et al, 2017). The advent of Whole Exome Sequencing (WES) has led to the discovery of specific gene mutations in clones or subclones of CLL patients, affecting prognosis, disease progression and therapeutic response (Landau et al, 2015). TP53 (tumour protein p53) gene mutations, seen in about 10% of patients at initial diagnosis, are commonly associated with del17p and lead to an aggressive, often refractory, disease course with poor therapeutic response, due to a TP53-mutated clone selection (Rossi et al, 2014).
TP53 mutations can be found in healthy older populations and were detected in some patients with acute myeloid leukaemia (AML)/myelodysplastic syndrome before chemotherapy (Wong et al, 2015). Furthermore, mice bearing both normal and mutated clones, expand the latter after chemotherapy (Wong et al, 2015). These findings suggest that TP53 mutation could precede tumour occurrence, although formal evidence in humans is lacking, due to the absence of adequate pre-tumoural samples.
Using deep Next Generation Sequencing (NGS; Appendix S1), we evaluated TP53 status in a patient, whose family included 3 generations of males (n = 5) affected with CLL who was diagnosed with B cell CLL, stage 0/A when aged 67 years. Analysis was performed longitudinally from 16 months before CLL manifestation to 30 days after a first-line ineffective treatment with rituximab plus bendamustine (R-Benda). At 16 months before clinical diagnosis his white blood cell (WBC) count was normal (7·9 × 109/l; 51·5% neutrophils, 40·5% lymphocytes), but was 83 × 109/l (neutrophil count 0·3 × 109/l) at 30 days post-chemotherapy. A TP53 gene mutation (c.548C>G, p.Ser183*), previously reported as pathogenic (Dicker et al, 2009), was detected in both pre- and post-diagnosis samples with 15% mutation-positive DNA fragments in the pre-diagnosis sample compared to 93·8% in the post-chemotherapy sample (Fig 1). The same mutation was absent in another affected family member, in line with a somatic event rather than a constitutive molecular alteration. The identified mutation could probably be the expression of a pre-existing monoclonal B-cell lymphocytosis and would account for the aggressive disease course (Ojha et al, 2014). At diagnosis, Fluorescence In Situ Hybridisation (FISH) analysis on peripheral blood cells showed deletion of chromosome 11 and 13 in 80% and 45% of the analysed nuclei, respectively (Fig 1) along with normal configurations of chromosome 12 and 17, which persisted over time. The IGHV gene was un-mutated.
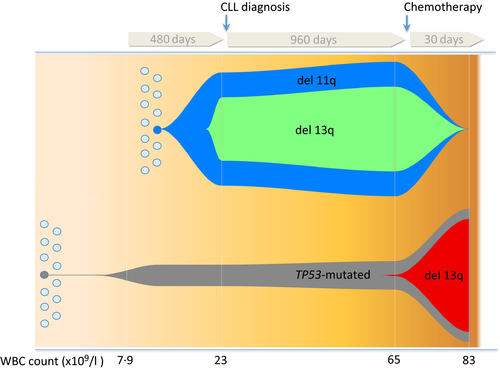
Genomic analysis by array-comparative genomic hybridization unmasked a complex picture evolving over time (Fig 1). No chromosomal abnormalities were detected 480 days before diagnosis. The initial clone with the 11q deletion and its subclone with the 13q deletion were undetectable after 30 days of therapy. Thus, a new TP53 subclone harbouring a 16·8 Mb type 2 deletion in chromosome 13, including DLEU1/2 together with the RB1 gene, arose and expanded up to 80% (Parker et al, 2011) (Fig 1). Results indicate that the TP53-bearing cell population was derived from different progenitor cells as a result of an earlier event preceding the del11- clone (80%) and del13- subclone (45%) responsible for the initial clonal expansion leading to CLL diagnosis. These findings expand the “oligoclonal theory” model, based on the evidence of the independent generation of TP53 mutated clones in AML (Griffith et al, 2015) demonstrating that somatic TP53 mutation precedes clonal expansion by years and, thus, clinical/laboratory diagnosis of CLL. Indeed, 16 months (480 days) before CLL diagnosis, the TP53 mutation load was 15%, meaning that 30% of cells were already bearing a heterozygous TP53 mutation.
The TP53-mutated clone was positively selected and consolidated during chemotherapy in accordance with previous evidence suggesting TP53 as the main gene responsible for relapse (Wong et al, 2015). Moreover, surpassing this pre-existent evidence, the present case provides evidence that a low-level clonal TP53 mutation not only acts as a “driver” of tumour development and progression but, notably, it antecedes cancer onset, acting as pre-clinical warning site of the tumour occurrence and as a very early-stage biomarker of a CLL disorder. This finding is in line with previous data, which found that more than 2% of individuals harbour a very small fraction of blood cells containing mutations that may represent premalignant events and probably define the earliest stages of a clonal haematopoietic expansion (Xie et al, 2014). Although CLL affects functionally inert mature B cells, we cannot exclude that the TP53 mutation greatly antecedes the clonal expansion that could even lie in stem cell progenitors.
Knittel et al (2017) demonstrated that the allelic frequency of the TP53 mutation increased, from very low to high, in two CLL patients after fludarabine/cyclophosphamide chemotherapy, suggesting that genotoxic therapy can select this aggressive clone. We underline the importance of avoiding genotoxic therapy in patients bearing TP53 mutation, even at very low mutational load. Indeed, our patient is currently on therapy with ibrutinib (420 mg/day), being un-responsive to first-line therapy with R–Benda and a second-line treatment with rituximab plus low-dose fludarabine, cyclophosphamide (FCR lite) and granulocyte colony-stimulating factor. Thus, given the positive familial history for lymphocytic tumours, a pre-clinical stage NGS TP53 analysis on a blood sample would have ranked our patient as a high-risk, low-responding individual, leading us to promptly opt for the latest generation targeted therapies available. A very low TP53 mutational load can be detected only by NGS, which should be included in current clinical practice to ensure personalised therapy and the best clinical management. Although current guidelines recommend genetic tests (FISH, IGHV mutational status, TP53 mutational status) when clinically indicated (relapse, refractory disease) or locally available, they can undoubtedly be useful to identify high risk patients. Interestingly, restoration of TP53 expression by a Cre-LoxP system in a mouse model was shown to be curative, leading to the selective regression of autochthonous lymphomas and sarcomas (Ventura et al, 2007). In the future, early-stage personalized gene therapy, tailored to ‘snip out’ the patient's specific TP53 mutation and replacing it with the wild type allele or a killer gene, could be of interest for CLL, bypassing the risk of a TP53-mutated subclone selection.
Acknowledgements
The Cell lines and DNA bank of Rett Syndrome, X-linked mental retardation and other genetic diseases, member of the Telethon Network of Genetic Biobanks (project no. GTB12001), funded by Telethon Italy, and the EuroBioBank network, provided us with specimens. We thank the patient and the Italian Leukaemia Association (Siena section) for their cooperation. We thank Dr. Donatella Raspadori for help revising the paper.
Ethics statement
This study was consistent with Institutional guidelines and approved by the ethical committees of Azienda Ospedaliera Senese, Siena. Informed consent was obtained from the patient.
Authors' contributions
A.M.P, F.T.P, M.B, A.G, took care of the clinical part of the study and wrote the paper, A.R. designed the research strategy, analysed the data and wrote the paper, E.F. analysed the data and wrote the paper, I.M. performed experiments and analysed the data, R.T, R.C. and C.F. performed experiments, L.D. and M.A.M. performed genetic counselling and provided patient samples, F.C. wrote the paper.
Disclosure of conflicts of interest
None.




