Applications of spatial transcriptomics in studying spermatogenesis
Abstract
Spermatogenesis is a complex differentiation process that is facilitated by a series of cellular and molecular events. High-throughput genomics approaches, such as single-cell RNA sequencing, have begun to enable the systematic characterization of these events. However, the loss of tissue context because of tissue disassociations in the single-cell isolation protocols limits our ability to understand the regulation of spermatogenesis and how defects in spermatogenesis lead to infertility. The recent advancement of spatial transcriptomics technologies enables the studying of the molecular signatures of various cell types and their interactions in the native tissue context. In this review, we discuss how spatial transcriptomics has been leveraged to identify spatially variable genes, characterize cellular neighborhood, delineate cell‒cell communications, and detect molecular changes under pathological conditions in the mammalian testis. We believe that spatial transcriptomics, along with other emerging spatially resolved omics assays, can be utilized to further our understanding of the underlying causes of male infertility, and to facilitate the development of new treatment approaches.
1 INTRODUCTION
Spermatogenesis is a fundamental biological process through which the spermatozoa are produced. This process starts with the proliferation and differentiation of spermatogonial stem cells (SSCs), which then undergo a series of mitotic and meiotic divisions, ultimately leading to the formation of spermatozoa. The developmental stages of spermatogenesis occur within the seminiferous tubules, where different cell types maintain a highly organized spatial arrangement. For example, SSCs, a specific subset of undifferentiated spermatogonia (SPG), reside near the basal membrane of the seminiferous tubules and form the basis of spermatogenesis and male fertility. These cells maintain a balance between self-renewal and differentiation,1, 2 which is closely regulated by a specialized microenvironment referred to as the “niche.”3-7 The SSC niche comprises multiple somatic cell populations, including Sertoli cells, peritubular myoid cells, Leydig cells, macrophage, innate lymphoid cells, mesenchymal cells, and endothelial cells, which work together to support SSC function and spermatogenesis.5, 8-11 Once differentiated, SPGs undergo successive mitotic divisions to replenish the germ cell pool. As germ cells progress through spermatogenesis, they migrate toward the lumen of seminiferous tubules. During this migration, germ cells undergo meiosis to produce haploid spermatids, accompanied by DNA methylation, chromatin remodeling, and significant morphological changes, ultimately leading to the continuous production of spermatozoa.1, 12-17
Accompanying the stereotypical spatial localization of different spermatogenetic cell types are the spatially restricted cellular and molecular events. For instance, the seminiferous tubules have been known to be protected by a unique blood‒tissue barrier known as the blood‒testis barrier (BTB).18, 19 The BTB locates near the basement membrane and segregates the seminiferous epithelium into the basal and the adluminal (apical) compartment. All the events related to meiosis I/II and post-meiotic spermatid development take place behind the BTB in the adluminal compartment. While the BTB is one of the tightest blood‒tissue barriers, it undergoes extensive remodeling during the seminiferous epithelial cycle in particular at stage VIII (mouse and rat) when preleptotene spermatocytes are being transported across the BTB to enter the adluminal compartment to prepare for meiosis I/II. Studies have shown that several Sertoli cell-expressing proteins are spatiotemporally regulated to facilitate the BTB remodeling process such as microtubule affinity-regulating kinase 4 (Mark4),20 Formin 1,21 and actin-related protein 3 (Arp3).22 Similarly, apical tubulobulbar complexes (apical TBCs; also known as apical ectoplasmic reticulum) are actin-based structures that help establish close contact between Sertoli cell-elongated spermatids near the lumen of seminiferous tubules. Apical TBCs undergo disassembly to facilitate spermatid release from the seminiferous epithelium at stage VIII of the mouse and rat cycle of the seminiferous epithelium.23-25 Studies have shown that adhesion molecules such as Nectin 2, Nectin 3, and α6 Integrin26 as well as actin regulatory proteins such as Arp322 and epidermal growth factor receptor pathway substrate 8 (Eps8)27 are concentrated at the sites of apical TBCs to facilitate their assembly and disassembly.
While pioneering studies such as those described above have provided a wealth of information on the cellular and molecular events associated with spermatogenesis, our understanding of these events is still incomplete especially in the context of human spermatogenesis because of ethical constraints and a lack of high-throughput tools to survey all the molecular interactions in the human testis. The emergence of single-cell sequencing technologies, combined with multi-omics approaches, has provided new insights into transcriptional and epigenetic regulations across the germ cell differentiation process.9, 11, 28-34 For example, through single-cell RNA sequencing (scRNA-seq), the germ cell developmental trajectory can be resolved, and various previously underappreciated cell states were discovered.9, 35 Despite these significant insights, scRNA-seq has limitations in revealing the molecular basis of spermatogenesis within the intact tissue context. During the tissue dissociation procedure of the scRNA-seq sequencing library preparation, critical information about cell‒cell interactions, cell location, and morphologies is lost.36, 37
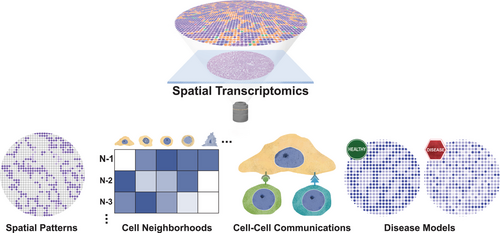
The past decade has witnessed remarkable advances in spatial transcriptomics (ST) technologies, driven by innovations in oligonucleotide synthesis, single-cell sequencing, fluorescent microscopy, and computational methods.38-43 These technologies have revolutionized our ability to study gene expression within the native tissue context, offering unprecedented insights into complex biological processes such as spermatogenesis. To date, multiple ST approaches have been applied to study mammalian spermatogenesis.33, 34, 44-46 In the following sections, we provide an overview of ST technologies, discuss how ST technologies facilitate our understanding of the complex cellular and molecular dynamics of mammalian spermatogenesis, and how these insights offer potentials for improving clinical diagnoses and treatment of male infertility (Figure 1).
2 OVERVIEW OF ST TECHNOLOGIES
Based on their experimental principles, ST techniques can be broadly classified into the following major branches: (1) region of interest (ROI) selection, (2) in situ sequencing (ISS), (3) in situ hybridization (ISH), and (4) solid-phase capture (Table 1).
| Year | Methods | Features | Number of targets | Spatial resolution | Limitations |
|---|---|---|---|---|---|
| ROI selection | |||||
| 1996 | LCM | Laser-based isolation of target cells | Entire transcriptome | Single cell | Low throughput and time-consuming |
| 2017 | Geo-seq | LCM combined with scRNA-seq | Entire transcriptome | Single cell | Low throughput and complex protocols |
| 2021 | STRP-seq | Imaging-free ST using tissue strips and computational reconstruction | Entire transcriptome | Strip-based resolution | Requires precise tissue cutting and computational reconstruction |
| 2023 | Laser-Seq | Combines laser microdissection with scRNA-seq | Entire transcriptome | Single cell | Low throughput and requires precise laser control |
| ISS | |||||
| 2013 | ISS | Padlock probes, RCA, and sequencing-by-ligation | 256 | Single cell | Low efficiency and laborious protocol |
| 2014 | FISSEQ | Reverse transcription with random hexamers | Entire transcriptome | Subcellular | Low sequencing depth and time consuming |
| 2018 | STARmap | Snail probes | 1000 | Single cell | High cost and complex protocol |
| 2023 | STARmap-X | Enhanced STARmap with higher multiplexing and sensitivity | >10,000 | Single cell | High cost and complex protocol |
| 2020 | HybISS | Padlock probes and RCA | 1000+ | Single cell | Complex imaging and sequencing protocol |
| 2021 | ExSeq | Hydrogel expansion and ISS | Entire transcriptome | Subcellular | Complex protocol |
| 2022 | BARseq2 | Barcoded ISS with improved throughput and sensitivity | Entire transcriptome | Single cell | Complex library preparation and sequencing pipeline |
| ISH | |||||
| 1998 | smFISH | Multiple oligonucleotide probes hybridize to the same transcript | Single target | Subcellular | Limited multiplexing because of spectral overlap |
| 2014 | seqFISH | Sequential rounds of hybridization, imaging, and probe stripping | >10,000 | Single cell | Time consuming and costly |
| 2017 | seqFISH+ | Enhanced version of seqFISH with higher multiplexing and error correction | >10,000 | Single cell | Complex imaging and analysis pipelines |
| 2015 | MERFISH | Binary encoding using modified Hamming codes | ∼10,000 | Single cell | Requires long transcripts (>3 kb) and complex equipment |
| 2016 | ExFISH | Combines hydrogel expansion with smFISH for enhanced resolution | <10 | Subcellular | Complex protocol and low throughput |
| Solid-phase capture | |||||
| 2016 | ST | Spatial barcodes and NGS | Entire transcriptome | ∼100 µm | Low sensitivity for rare transcripts and low spatial resolution |
| 2019 | Visium | Commercialized ST method | Entire transcriptome | 55 µm | Low spatial resolution |
| 2019 | Slide-seq | cDNA capture on 10 µm beads | Entire transcriptome | 10 µm | Low sensitivity for lowly expressed genes |
| 2021 | Slide-seqV2 | Improved bead chemistry and capture efficiency | Entire transcriptome | 10 µm | Not true single-cell resolution |
| 2020 | DBiT-seq | Microfluidic barcoding and NGS | Entire transcriptome | 10 µm | Requires specialized equipment |
| 2021 | Stereo-seq | DNA nanoball arrays | Entire transcriptome | 220 nm | High cost |
| 2021 | Seq-Scope | Repurposes Illumina flow cells | Entire transcriptome | Subcellular | Complex protocol |
| 2021 | PIXEL-Seq | Scalable polony gel stamping for array fabrication | Entire transcriptome | Subcellular | Complex protocol |
| 2022 | HDST+ | High-density ST with improved barcoding efficiency | Entire transcriptome | 2 µm | Requires advanced imaging and computational analysis |
| 2023 | Visium HD | 10× genomics’ next-generation platform with subcellular resolution | Entire transcriptome | Subcellular | High cost |
- Abbreviations: ExFISH, expansion fluorescence in situ hybridization; ExSeq, expansion sequencing; FISSEQ, fluorescent in situ RNA sequencing; HybISS, hybridization-based in situ sequencing; ISH, in situ hybridization; ISS, in situ sequencing; LCM, laser capture microdissection; MERFISH, multiplexed error-robust fluorescence in situ hybridization; RCA, rolling circle amplification; ROI, region of interest; scRNA-seq, single-cell RNA sequencing; seqFISH, sequential fluorescence in situ hybridization; smFISH, single-molecule fluorescence in situ hybridization; NGS, next generation sequencing.
2.1 ROI selection
ROI selection-based ST is to select the tissue regions of interest for downstream sequencing and analysis. Geo-seq,47 Laser-Seq,48 and STRP-Seq49 use laser capture microdissection (LCM)50, 51 to dissect ROIs, followed by RNA sequencing (RNA-seq) or scRNA-seq. These methods enable transcriptomic analysis of serial tissue slices, which can then be reconstructed to map spatial gene expression. ROI selection can be also achieved through photoconversion techniques, such as NICHE-Seq,52 which uses photocaged fluorescent reporters to label specific areas. Similarly, PHOTON uses photocaged fluorescent oligos to allow selective sequencing of RNA species at a subcellular resolution.53 Collectively, ROI selection-based ST technologies allow researchers to isolate and analyze specific cells or regions within heterogeneous tissue, ensuring that only the target areas are analyzed in precise locations from tissue context. This also means that these approaches are not suitable for unbiased whole tissue ST.
2.2 In situ sequencing
ISS-based ST approaches rely on imaging-based fluorescent readout of mRNA barcodes. In these approaches, barcoded padlock probes are often used to hybridize to specific RNA molecules. Following ligation of the padlock probes to form a circular structure, rolling circle amplification is performed in situ to generate thousands of copies of the same mRNA barcodes to allow sensitive detection of the barcodes through rounds of in situ imaging.54, 55 To further increase the spatial resolution and the throughput of ISS, expansion sequencing (ExSeq)56 was developed to combine hydrogel expansion method to physically enlarge the specimens with ISS. Although ISS-based ST approaches achieve subcellular resolution, these methods require complex workflows and extensive image analysis expertise, making them challenging to be implemented in regular laboratories.
2.3 In situ hybridization
Single-molecule fluorescence in situ hybridization (FISH) was developed back in the 90s, allowing for the precise localization and quantification of individual RNA molecules within cells. Aiming for higher throughput of RNA detection, highly multiplexed fluorescent probe hybridization of RNA transcripts followed by imaging has been developed. Techniques such as multiplexed error-robust FISH57, 58 and sequential FISH59, 60 now allow spatial readout of thousands of gene transcripts at subcellular resolution. However, like ISS, ISH-based ST approaches present a high technical barrier for non-experts.
2.4 Solid-phase capture
Most solid-phase capture-based ST methods work by casting a thin tissue slice onto a pre-indexed array for spatial capturing and indexing of mRNA molecules from the tissue. Built upon the work of Stahl et al.61 to capture poly-adenylated RNA molecules on a spatially barcoded microarray with unique molecule indices, 10× Genomics Visium61 was developed with 55 µm in spot diameter and 200 µm in center-to-center distance between spots. Later on, Slide-seq62, 63 achieves a spot resolution of 10 µm by using spatially indexed beads. In recent years, new technologies have emerged. For example, DBiT-seq64 uses microfluidic barcoding for high-resolution spatial mapping, and Stereo-seq65 employs DNA nanoball arrays to achieve a spatial resolution of 220 nm. Seq-Scope66 and PIXEL-Seq67 have pushed the cutting-edge further by offering subcellular resolution and scalable array fabrication, respectively. These methods have become versatile tools for studying spatial gene expression in complex tissues.
3 ST REVEALS THE SPATIAL GENE EXPRESSION PATTERNS IN THE TESTIS
Genes with non-randomly distributed expression patterns may be of significance to tissue and cell functions. Thus, a systematic identification of these genes may lay a solid foundation for functional discoveries. By applying SPARK68—a statistical framework that fits the spatial count data through generalized linear spatial models—to the Slide-seqV2 data of the mouse testis, Chen et al. identified a set of spatially patterned (SP) genes in individual seminiferous tubules.45 Since seminiferous tubules can be classified into different stages along the cycle of the seminiferous epithelium, the stage-specific SP genes were further identified.60 Among the SP genes, Habp4, whose function was largely elusive, was shown to be associated with chromatin remodeling process during spermiogenesis.44 Besides SP genes within the seminiferous tubules, ST also facilitated the identification of SP genes within somatic cells in the interstitial space of the testis. For example, 1700017N19Rik was found to be enriched in Leydig cells that were spatially adjacent to stage IV–VI seminiferous tubules.44 In addition, by applying RNA SeqFISH+ to profile 2638 genes in mouse testis cross-sections, Chakravorty et al. identified a periodic transcriptional pattern in Sertoli cells that was closely synchronized with germ cell development while other somatic cells do not demonstrate such synchronization.46 Such a cyclic transcriptional feature in Sertoli cells, including the one associated with the retinoic acid signaling, remains largely unperturbed even after the germ cells are ablated with busulfan,62 suggesting a Sertoli cell-autonomous transcription program at play.
In addition to the adult mammalian testis, ST has also been applied to study gonadal development in the mouse and the human. One study leveraged scRNA-seq to identify a previously uncharacterized somatic cell population that is PAX8 positive.33 ST data suggest that this population locates at the gonadal‒mesonephric border where the rete testis develops in the male testis from late postconceptional weeks. This cell population shows a unique transcriptional pattern of axon guidance factors, suggesting that they have a structural and supporting role for rete testis development.33
4 ST ENABLES THE CHARACTERIZATION OF CELL NEIGHBORHOODS
Spatial co-localization of cells often indicates functional interactions. Chen et al. calculated the spatial proximity between distinct testicular cell types using the Slide-seqV2 data to determine spatial contact frequencies between cell types.44 This was performed through a K-nearest neighbor approach to count the frequency of each cell type within the spatial neighbors of a given cell type. Through this approach, they discovered that the cellular composition of the microenvironment surrounding SPGs—a group of cells characterized by dynamic cell state changes and a delicate balance between self-renewal and differentiation—exhibited notable differences between the human and the mouse.44 Their findings revealed that in humans, the spatial composition of cell types surrounding the undifferentiated SPGs differs from that surrounding the differentiating SPGs, whereas no such difference was observed in the mouse testis.44 This observation suggests that different mechanisms may exist to regulate SPG activities between the mouse and the human.
5 ST ENABLES THE CHARACTERIZATION OF CELL‒CELL COMMUNICATIONS
The activities of SSCs are tightly regulated by their surrounding microenvironment. Rajachandran et al. systematically characterized the interactions between the SSCs and their surrounding cells through calculating the strength of ligand‒receptor (LR) interactions by multiplying the gene expression levels of the ligands from the sender cells with the expression levels of corresponding receptors from the neighboring receiver cells using the Slide-seqV2 data.45 Through this analysis, they revealed numerous potential niche factors involved in regulating SSCs activities. For example, in the mouse testis, they found that pleiotrophin (PTN)—a ligand secreted by multiple cell types including the Leydig cells and endothelial cells—influenced SSC activities partially through syndecan (SDC) receptors.45 They then confirmed this observation through SSC transplantation assays using SPGs treated with PTN. They found that cultured SPGs treated with PTN at 5 ng/mL resulted in an increased colony size after being transplanted into the testes of donor mice deprived of endogenous germ cells when compared with untreated control SPGs.45 This was likely because of the ability of PTN to increase the number of SPGs with colonization potential. In addition, using the same computational approach, they identified ephrin A1 (EFNA1), produced mostly by endothelial cells, as a potential niche factor for SSC function in the human testis.45 However, the functional role of EFNA1 on human SSCs remains to be confirmed by follow-up experiments such as the xenotransplantation assay.
Rajachandran et al. also identified LR pairs with stage-specific activities in the seminiferous tubules,45 indicating that distinct signaling pathways are at play at different stages of the cycle of the seminiferous epithelium. Moreover, they leveraged the testis Slide-seqV2 data to enable the inference of signaling directions across the testis tissue sections using a computational tool called COMMOT.69 COMMOT infers spatial signaling directions by interpolating the cell-by-cell cell‒cell communication matrix to a vector field to identify the direction from which the signal is received or sent. Through cell type-specific activity analysis, identified LR pair-mediated signaling pathways differentially received by different testicular cell types in humans were also identified. For example, Rajachandran et al. revealed that the fibroblast growth factor signaling pathway primarily effects a subset of SPGs, while myoid cells are the major cell type to receive the signaling activities of both platelet-derived growth factor pathway and Visfatin pathway.45
6 ST FACILITATES THE DETECTION OF MOLECULAR CHANGES IN TESTES FROM DISEASE MODELS
ST assays have been applied to capture molecular changes in the testis under pathological conditions.45 By applying Slide-seqV2 to testes from leptin-deficient diabetic mice, Chen et al. found that multiple genes such as Smcp, Odf1, and Malat1 showed altered spatial expression patterns in diabetic testis cross-sections when compared with those from wild-type (WT) mice. At the cellular level, by calculating pairwise spatial contact frequencies between cell types, the authors showed that diabetic mouse testes manifested significant disruptions in the spatial gene expression patterns in seminiferous tubules compared with the WT. For example, Smcp, a gene that was spatially enriched at the luminal region of a WT seminiferous tubule, was found to be diffusively expressed throughout the seminiferous tubules of diabetic mice. This change in spatial gene expression pattern might be partially explained by a change in cellular distribution in the seminiferous tubule.44 For example, compared to WT seminiferous tubules where elongating spermatids (ESs) cluster together near the lumen, ESs were found away from the seminiferous tubule lumens and were more likely to make spatial contacts with other cell types.44 This cellular change was accompanied by alterations in the spatial expression patterns of a subset of LR pairs. Functional enrichment analysis showed that these LR pairs with altered spatial expression patterns were associated with immune cell functions, indicating that the inflammatory response might underlie diabetes-induced testicular injuries.45
ST analysis has also been applied to study human seminoma, a common malignant tumor in the male testis.34 A high similarity in the transcriptome between seminoma and primordial germ cells (PGCs) was observed. For example, seminoma cells and PGCs shared the expression of key transcription factors such as TFAP2C and SOX17, reinforcing the notion that seminoma may originate from PGC-germ cell neoplasia in situ. Further analysis revealed a complex immune‒tumor interaction within the seminoma tumor microenvironment (TME). For example, CD4+ and CD8+ T-cell populations surrounding the seminoma exhibited features of dysfunction or exhaustion, suggesting an immune-suppressed state that may contribute to tumor progression.34 Additionally, macrophage migration inhibitory factor was found to be highly expressed in the seminoma samples, and its receptor was mainly expressed on the surrounding immune cell populations.34 This indicates frequent crosstalk between the tumor cells and the immune cells in the TME, which could potentially be targeted for therapeutic purposes.
7 DISCUSSION
The integration of ST into male fertility research holds immense promise in unraveling the complex regulatory networks underlying male fertility. By capturing the spatial context of gene expression within the native environment of the testis, ST enables the identification of key genes and pathways critical to the regulation of spermatogenesis. In addition, ST methodologies facilitate the probing of germ line‒soma interactions within the testicular microenvironment, providing insights for advancing in vitro intervention and reconstitution of germ cells. Moreover, comparative spatial analyses across species offer valuable insights into the evolutionary dynamics of reproductive biology.
The discoveries from analyses of the mouse and human testis ST data reviewed in this manuscript represent only a tip of the iceberg of what can be understood about mammalian spermatogenesis. More insights can be gained through further mining existing ST data as more computational tools are being developed. For instance, besides identifying LR interactions between testicular cell types, further analyses can be performed to identify downstream activated transcription factors (TFs) by the LR interactions and target genes of the TFs.70 Furthermore, a cell's surrounding microenvironment affects its gene expression profile. To this end, analyses can be performed to identify SSC-specific niche-associated genes, defined as SSC genes whose expression are significantly up- or down-regulated in the context of specific spatial niches during the cycle of the seminiferous epithelium.71 Finally, as more ST data being generated and are deposited into publicly available data visualization portals (see testis Slide-seqV2 data in the single-cell portal as an example: https://singlecell.broadinstitute.org/single_cell/study/SCP2166/testis-slide-seq), researchers with limited computational expertise can now begin to survey their genes/cell types of interests using those ST data.
Looking ahead, improvement of ST technologies will further our mechanistic understanding of the regulation of male fertility.
First, the refinement of the spatial resolution of array-based ST approaches would help to elucidate the intricate molecular interactions between testicular cell types. Previous work using array-based ST approaches suffer from the issue of the mixing of RNA from multiple cells captured by individual spots on the array. Although RNA de-mixing methods, such as robust cell-type decomposition,72 have been developed to address this issue, these methods rely on a matching scRNA-seq reference and therefore cannot resolve cell types or cell states that have not been identified by scRNA-seq. More recent technologies such as Stereo-seq65 and Slide-tags73 may be applied to testicular tissues thanks to their improved spatial resolution. Other ISS and ISH-based approaches such as ExSeq56 and SeqFISH+60 may also serve as good alternatives as they can achieve ST profiling at the subcellular level. However, testis-specific image analysis pipelines need to be developed to handle the challenging task of image registration, segmentation, and transcript quantification.
Second, expanding the abilities of the spatial technologies to capture information beyond the transcriptome would provide new opportunities for mechanistic discoveries. For instance, the non-coding and non-polyadenylated RNA species (e.g., small RNAs) play important roles in testicular functions.74, 75 However, most of the array-based ST methods are limited in detecting only mRNAs as these methods rely on capturing the poly-A tails of RNAs. Thus, combining total RNA-seq with spatial readout would reveal novel insights that have been previously overlooked. Furthermore, combining spatial readout with epi-transcriptome (such as RNA methylation) profiling using Oxford Nanopore Technologies, as well as with nascent RNA analysis methods such as global run-on sequencing76 (GRO-Seq) or native elongating transcript sequencing77 (NET-Seq), could provide more detailed insights into RNA dynamics and modifications.78, 79 Besides incorporating various RNA-based modalities, the integration of ST with DNA-based modalities such as the spatial epigenome and the three-dimensional genome structure would propel new discoveries of gene regulatory mechanisms in the testis. Several research groups have recently developed approaches for spatially co-profiling the epigenome and transcriptome. For example, spatial ATAC/CUT&Tag-RNA-seq, a technology built upon the spatial barcoding strategy first implemented in DBiT-seq,64 enables the simultaneous detection of chromatin accessibility status, protein‒chromatin interactions, and the transcriptome within intact tissue sections.80 The Slide-tags technology also enables the co-capture of the spatially resolved chromatin accessibility and the transcriptome at single-cell resolution.73 In addition, the emerging spatial metabolomics and spatial proteomics technologies will provide opportunities to study metabolic and post-translational regulation during spermatogenesis.81-84 Finally, innovative approaches such as Perturb-map, which integrates ST with in situ CRISPR screens, will enable the identification of genetic determinants or regulatory mechanisms underlying various biological processes.85, 86
Third, addressing the cost barriers associated with ST experiments is also crucial for broadening their accessibility and scalability. Technologies with simplified workflow would also democratize access to ST tools for a wider community.
In conclusion, the male fertility research will continue benefiting from the increased levels of spatial resolution, yield, and accessibility of ST technologies. As spatial technologies are under rapid development and refinement, we are poised to unlock new insights into the mechanisms that regulate fertility.
AUTHOR CONTRIBUTIONS
Qianlan Xu and Haiqi Chen performed literature review and wrote the manuscript.
ACKNOWLEDGMENTS
This work was supported by an NIH/NICHD grant to H.C. (R01HD114698).
Open Research
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.




