The human sperm proteome—Toward a panel for male fertility testing
Abstract
Background
Although male factor accounts for 40%–50% of unintended childlessness, we are far from fully understanding the detailed causes. Usually, affected men cannot even be provided with a molecular diagnosis.
Objectives
We aimed at a higher resolution of the human sperm proteome for better understanding of the molecular causes of male infertility. We were particularly interested in why reduced sperm count decreases fertility despite many normal-looking spermatozoa and which proteins might be involved.
Material and methods
Applying mass spectrometry analysis, we qualitatively and quantitatively examined the proteomic profiles of spermatozoa from 76 men differing in fertility. Infertile men had abnormal semen parameters and were involuntarily childless. Fertile subjects exhibited normozoospermia and had fathered children without medical assistance.
Results
We discovered proteins from about 7000 coding genes in the human sperm proteome. These were mainly known for involvements in cellular motility, response to stimuli, adhesion, and reproduction. Numbers of sperm proteins showing at least threefold deviating abundances increased from oligozoospermia (N = 153) and oligoasthenozoospermia (N = 154) to oligoasthenoteratozoospermia (N = 368). Deregulated sperm proteins primarily engaged in flagellar assembly and sperm motility, fertilization, and male gametogenesis. Most of these participated in a larger network of male infertility genes and proteins.
Discussion
We expose 31 sperm proteins displaying deviant abundances under infertility, which already were known before to have fertility relevance, including ACTL9, CCIN, CFAP47, CFAP65, CFAP251 (WDR66), DNAH1, and SPEM1. We propose 18 additional sperm proteins with at least eightfold differential abundance for further testing of their diagnostic potential, such as C2orf16, CYLC1, SPATA31E1, SPATA31D1, SPATA48, EFHB (CFAP21), and FAM161A.
Conclusion
Our results shed light on the molecular background of the dysfunctionality of the fewer spermatozoa produced in oligozoospermia and syndromes including it. The male infertility network presented may prove useful in further elucidating the molecular mechanism of male infertility.
1 INTRODUCTION
An estimated 8%–15% of couples are involuntarily childless worldwide, with male factor infertility accounting for 40%–50% of the cases.1-3 Although the causes of the disorder are often suspected in reduced sperm quantity and/or quality, the proteome of the spermatozoon has not yet been conclusively elucidated. Instead of giving the full picture, the available data appear to mainly reflect the rising resolution of the analytical pipelines employed. For example, while the liquid chromatography–mass spectrometry (MS) study by Baker et al.4 detected 1056 gene products in human spermatozoa, the pipeline employed by Wang et al.5 enabled the determination of 4675 sperm proteins. Larger numbers of 6198 and 6238 human sperm proteins were gathered in subsequent reviews based on proteomics studies.6, 7 The recent compilation by Castillo et al.8 even contained 6871 human sperm proteins. However, such a scale has not been confirmed in single MS analysis so far.
As with the sperm proteome, the molecular causes of male fertility impairment are only partially understood. Previous studies in this area were based on transcript abundances in testes and spermatozoa9-11 or consulted a methodologically broad range of published data.12, 13 In addition, mutations, copy-number variations, and polymorphisms in the genome have been checked for associations with infertility.14, 15 Also, epigenetic deviations may be associated with fertility impairment16; however, this is still debated.17 Others derived fertility relevance from parameters such as sequence conservation and phenotypes of murine knockouts,18, 19 and quantitative proteomics of spermatozoa is increasingly being incorporated.20-22 Yet, the diversity of approaches has not made it easier to define a standard panel of male fertility markers for routine diagnostics. Rather, the corner stone of male fertility diagnostics remains to be the recording of spermiogram parameters.7, 23
For obvious reasons, the absence of any spermatozoa in ejaculates or azoospermia causes infertility, whereas the presence of a certain amount of fully functional spermatozoa (normozoospermia) suggests fertility. But the question is as to why a reduction in sperm count can be critical to fertilization success, if there are still many motile and morphologically normal spermatozoa.24 In fact, the criteria of oligozoospermia (O) (i.e., less than 39 million spermatozoa per ejaculate and less than 16 million spermatozoa per milliliter) still allow for the thousands of seemingly unimpaired spermatozoa per ejaculate. The same may be asked for asthenozoospermia, that is, a decrease in the proportion of progressively moving spermatozoa to less than 30%, and teratozoospermia, that is, a decline in the percentage of spermatozoa with normal morphology to less than 4%.3, 25, 26 This is contrasted by only 100–1000 functional sperm cells, which assumedly reach the egg cell in natural fertilization.27 A possible explanation is that spermatozoa that appear to be fully functional are actually dysfunctional. Consistent with this, poorer semen parameters are accompanied by reduced success rates of assisted reproduction techniques (ARTs) despite the use of spermatozoa considered promising.24 The cause of such a dysfunction would then have to be sought in the molecular field, which would blur the boundaries to idiopathic infertility or infertility without phenotypic evidence.1, 12, 23, 28 In fact, about 40% of the infertile men are idiopathic infertile,14 which underlines the high need for progress in this field of diagnostics.
The present study aims at an in-depth reconstruction of the human sperm proteome and an elucidation of the molecular causes of male fertility losses. We were especially interested as to why fecundity is reduced in O and syndromes including it, and which proteins might be useful biomarkers for assessing the fertility status of respective men. To address these points, we carried out MS analysis of spermatozoa in an unprecedented sample of 76 men. Donors were either fertile and presented with normozoospermia or infertile and diagnosed with O, oligoasthenozoospermia (OA), or oligoasthenoteratozoospermia (OAT). In contrast to previous proteomic studies based on pools of sperm samples,21, 28-30 spermatozoa of single subjects were analyzed separately. From the differentially abundant proteins, we distilled out those, which should have elevated diagnostic potential. Finally, we demonstrate that non-overlapping sets of (candidate) markers of male fertility together establish the male infertility network.
2 MATERIALS AND METHODS
2.1 Study cohorts
Permission for sample collection was granted by the Ethics Committee of the Medical Faculty at the Martin Luther University Halle/Saale (approval number 218/14.04.10/2; date of decision: April 19, 2010; date of approved amendment: March 14, 2013). All participants were recruited from the fertility outpatient care of the University Hospital Halle (Saale) and gave written informed consent. Ejaculates were provided by 76 men (Table S1). Spermiogram parameters were re-assessed according to the updated version of the World Health Organization (WHO) laboratory manual for the examination and processing of human semen (6th edition, 2021),26 with no changes in the diagnoses of the study participants. For clarity reasons, we use descriptive terms for abnormal spermiograms (O, asthenozoospermia, teratozoospermia, and mixed forms) as proposed in previous WHO laboratory manuals (5th edition and earlier). Nonetheless, the authors acknowledge the fact that the cut-off values used for defining these terms do not necessarily determine the fertility status of an individual male. Thirty-eight infertile men (mean and median age = 32 years, each) presented with O (21), OA (7), or OAT (10), and were unintentionally childless, that is, their partners did not conceive despite regular unprotected intercourse within at least 12 months. In the 38 infertile patients, median sperm count per ejaculate was 36.5 mio (mean = 41.4 mio). The control cohort included 38 men (mean and median age = 30 years, each) presenting with normozoospermia, who already had fathered offspring without medical assistance. Median sperm count of the corresponding ejaculates was 155.0 mio (mean = 205.7 mio) (Table S1).
2.2 Sample preparation for MS
Upon swim-up, spermatozoa were separated from seminal plasma by centrifugation for 5 min at 1000 g. After discarding the plasma, cells were resuspended twice in phosphate-buffered saline and re-pelleted by centrifugation at 2000 g for 5 min, each time succeeded by discard of the supernatant. Washed spermatozoa were shock-frosted in liquid nitrogen and stored at –80°C. Following thawing on ice, samples were boiled in 1× lithium dodecyl sulfate buffer and separated by polyacrylamide gel electrophoresis on a 4%–12% gradient NOVEX gel (Thermo Scientific) for 10 min at 180 V. The gel was fixated and stained with Coomassie blue for 30 min and subsequently washed with water overnight. In-gel digestion was performed as previously described.31 In short, each sample was cut from the gel, minced, and transferred to a reaction tube. The gel pieces were destained with 50% EtOH/25 mm ammonium bicarbonate (ABC) buffer (pH 8.0). After dehydration of the gel pieces with pure acetonitrile (ACN), samples were dried for 5 min in a concentrator (Eppendorf) and afterward incubated with reduction buffer (10 mm dithiothreitol in 50 mm ABC) for 30 min at 56°C. The reduction buffer was removed, substituted with alkylation buffer (50 mm iodoacetamide in 50 mm ABC), and then subjected to 30 min incubation at room temperature in the dark. Gel pieces were completely dehydrated with pure ACN and covered in trypsin solution (1 µg trypsin in 50 mm ABC per sample). Proteins were digested overnight at 37°C. Tryptic peptides were extracted twice by incubation with extraction buffer (3% trifluoroacetic acid and 30% ACN) for 15 min and afterward with pure ACN. After concentration of the elution fraction to about 10%–20% in a concentrator (Eppendorf), the peptides were passed through a StageTip.32 StageTips were prepared using two layers of C18 material (Empore), which was activated with methanol, washed with solution B (80% ACN, 0.1% formic acid), and equilibrated once with solution A (50 mm ABC, 0.1% formic acid). Extracted peptides were loaded on the StageTips and washed with solution A. Peptides were eluted with 30 µL solution B, ACN was removed by use of a concentrator (Eppendorf), and samples were diluted with 6 µL solution A.
2.3 MS measurement
The samples were injected via an autosampler into an uHPLC (EASY-nLC 1000, Thermo Scientific). Peptides were loaded on a 25 cm capillary (75 µm inner diameter; New Objective) packed in-house with Reprosil C18-AQ 1.9 µm resin (Dr. Maisch) for reverse-phase chromatography. The EASY-nLC 1000 HPLC system was directly mounted to a Q Exactive Plus mass spectrometer (Thermo Scientific). Peptides were eluted from the column with a 208 min optimized gradient from 2% to 40% ACN with 0.1% formic acid at a flow rate of 225 nL/min with a column oven set-up operating at 40°C (Sonation). The heated capillary temperature was set to 250°C. Spray voltage ranged from 2.2 to 2.4 kV. The mass spectrometer was operated in data-dependent acquisition mode with one MS full scan and up to 10 triggered MS/MS scans using higher energy collisional dissociation. MS full scans were obtained in the orbitrap at 70,000 resolution with a maximal injection time of 20 ms, while MS/MS scan resolution was set to 17,500 and maximal injection for 120 ms. Unassigned and charge state 1 were excluded from MS/MS selection and peptide match was preferred. Detailed information can be obtained from the measurement files available on the public ProteomeXchange repository (PXD037531).
2.4 MS data analysis
Raw data analysis was performed with MaxQuant v1.5.2.833 with standard settings except label-free quantification (LFQ) and match between runs were activated. Time windows for matching and aligning were 0.7 and 20 min, respectively. MaxQuant analysis including detailed settings has been deposited at ProteomeXchange under the above accession number. After MaxQuant, potential contaminants, reverse database hits and proteins only identified by site were removed. Further, we filtered for a minimum of two identified peptides (minimum 1 unique). For quantitative proteome analyses, lacking values were imputed assuming a beta distribution within 0.2 and 2.5 percentiles of measured LFQ intensities per sample. Downstream comparisons of protein abundances focused on spermatozoa of infertile men diagnosed with reduced sperm count and fertile men with normal semen parameters. Differentially abundant sperm proteins were grouped according to infertility diagnoses, thus generating O list (O vs. normozoospermia), OA list (OA vs. normozoospermia), and OAT list (OAT vs. normozoospermia). For being contained in a list, a protein needed to be detected in at least two samples of a diagnosis, and abundances had to vary by a factor of three at minimum (false discovery rate [FDR] corrected p-value ≤ 0.05, Welch two sample t-test). This corresponds to a minimum difference of log2-transformed values of 1.585. We additionally highlight sperm proteins showing at least eightfold differential abundances, corresponding to a minimum difference of log2-transformed values of 3.000.
2.5 Further analyses
We ran ShinyGO v0.7634 on ENSEMBL stable IDs of the genes encoding MS-determined sperm proteins for deriving the lengths of coding sequences and transcripts. We also adopted from the ShinyGO output the grouping of genes according to higher level biological process gene ontologies (GOs). Moreover, we matched ENSEMBL stable gene IDs from present MS analysis with the genes to the sperm proteins collected by Castillo et al.8 For the latter compilation, UniProt/SwissProt IDs had been converted into ENSEMBL IDs using BioMart (Ensembl Genes 106, Human genes, GRCh38.p13).
Overlaps between above O-associated lists of genes/proteins were rendered in a Venn diagram (https://bioinformatics.psb.ugent.be/webtools/Venn/). We tested for the overrepresentation of biological process GOs in O, OA, and OAT lists using ClueGO v2.5.935 within Cytoscape v3.9.1.36 In addition, we matched the list for the most comprehensive infertility diagnosis, that is, the OAT list, with entries in NCBI's Online Mendelian Inheritance in Men (OMIM) database (state February 1, 2022) referencing male fertility impairment. Corresponding OMIM entries were related to O and non-obstructive azoospermia using all possible forms of spermatogenetic failure (SPGF) as search items: SPGF1, SPGF2, …, SPGF65, SPGFX1, SPGFX2, SPGFX3, SPGFY1, and SPGFY2. We further collected OMIM entries on asthenozoospermia, teratozoospermia, and male infertility. We screened the WWW for publications using the combination of symbol and infertil* as initial search items (e.g., APAF1 infertil*). We additionally queried The Human Protein Atlas v22 (http://proteinatlas.org/; accessed on January 10, 2023) for the expressional landscape of the genes behind differentially abundant sperm proteins.
Subsequently, we assessed if the candidate markers presented herein and additional ones emerging from external sources might be connected at a higher level. For this purpose, we reconstructed three protein–protein interaction (PPI) networks of growing complexity. Using standard settings in STRING v11.5,37 the first network was confined to the biomarkers as inferred from present empirical data. The second network additionally considered candidate markers as emerging from a previous computational approach. Corresponding genes/proteins were calculated to have elevated fertility probability (≥0.35) given enhanced sequence conservation, heightened testicular transcript abundance, and increased connectivity in a body-wide PPI network. Furthermore, knockouts of their murine orthologs associated with male fertility impairment.19 We extracted the aforementioned proteins from Dataset M at the PreFer Genes website (https://prefer-genes.uni-mainz.de/). A third network was reconstructed after further expansion of the sample by all proteins with previous mention in the context of male infertility in OMIM database (see above). The networks were compared by their overall coherence and average node degree, that is, the mean number of PPIs per protein or node. Each network was tested for PPI enrichment.
Raw p-values from chi-square tests, GO enrichment tests, and PPI enrichment tests were transformed into FDRs.38 Further consideration of test results required an FDR of 0.001 or lower.
3 RESULTS
3.1 Fine-scale reconstruction of the human sperm proteome
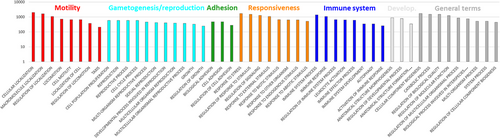
Present MS analysis identified 5778 protein groups (q ≤ 0.05, each) across sperm samples provided by 76 men and purified by swim-up (Table S1). These contained 33,191 gene products to which 6981 ENSEMBL stable gene IDs were available. Of the latter, 4545 had counterparts in the compilation of sperm proteins by Castillo et al.8 Thus, 65% of the sperm proteins for which we found evidence in human spermatozoa can be regarded as reproduced (Table S2). The 6939 genes mapped by ShinyGO significantly differed from the rest in the human genome by having shorter coding sequences (FDR = 6E-15) and transcripts (FDR = 1.7E-20; t-test, each) (Figure 1). A closer look for the 50 biological process GOs occurring with highest frequencies corroborated the importance of sperm motility for fertilization. Processes such as cell proliferation and reproduction, adhesion, and responsiveness to stimuli were also represented, as were involvements in developmental processes and the immune system. The latter included genes coding for spermatogenesis-associated protein 20 (SPATA20), zona pellucida-binding protein 1 (ZPBP), and sperm-associated antigen 4 (SPAG4) (Table S3). Additional higher ranking terms were of more general nature such as “catabolic process” (Figure 2). The GOs of the remaining 50 smaller protein groups partially confirmed roles in male gametogenesis (meiotic cell cycle process, spermatid nucleus differentiation) and fertilization (acrosome reaction), while others referred to additional entanglements (Table S3).

3.2 Exactness of MS analysis
For being considered, a protein had to be determined in at least one unique and one razor peptide across the 76 samples analyzed. However, much higher values were achieved in the validated and novel candidate markers detailed below (Tables 1–4 and S2). In case of the validated marker candidates, we had 4–246 (mean = 51, median = 32) determinations of unique and razor peptides. These were mostly unique peptides, as illustrated by corresponding mean and median numbers of 47 and 29, respectively. In the novel candidate markers, the span of unique and razor peptides ranged from 7 to 177 (mean = 33, median = 21). Again, these were mostly specific to a particular protein group, as indicated by mean (32) and median (21) values of unique peptides. We consider these counts as a confirmation of the reliability of the detections.
| Symbol | ID | OAT | O | OA | Functional annotation (UniProt) | THPA |
|---|---|---|---|---|---|---|
| ACTL9* | Q8TC94 | 0.173 | ↓ | ↓ | Acrosome biogenesis and perinuclear theca formation | I |
| CCIN* | Q13939 | 0.256 | ↓ | ↓ | Cytoskeletal element in spermiogenesis | I |
| CFAP47* | Q6ZTR5 | 0.128 | ↓ | ↓ | Flagellar formation and sperm motility | III |
| CFAP65* | Q6ZU64 | 0.236 | ↓ | ↓ | Flagellar formation and sperm motility | III |
| WDR66* | Q8TBY9 | 0.254 | ↓ | ↓ | Spermatozoa motility | III |
| DNAH1* | Q9P2D7 | 0.193 | ↓ | ↓ | ATPase activity; sperm flagellum motility; formation of the inner dynein arms and biogenesis of the axoneme | III |
| SPEM1* | Q5F289 | 0.261 | ↓ | ↓ | Cytoplasm removal during spermatogenesis | I |
| ACTL7A* | Q9Y615 | 0.238 | ↓ | Acrosome biogenesis | I | |
| MNS1* | Q9C512 | 0.133 | ↓ | Control of meiotic division and germ cell differentiation (…) during meiosis; sperm flagellum assembly | II | |
| PLCZ1* | Q86YW0 | 0.182 | ↓ | Initiates embryonic development | I | |
| PMFBP1* | Q8TBY8 | 0.270 | ↓ | Spermatogenesis (…); maintenance of sperm head and tail integrity | II | |
| QRICH2* | Q9H0J4 | 0.204 | ↓ | Scaffold protein (…); maintenance of sperm head and tail integrity | II | |
| SPAG17* | Q6Q759 | 0.184 | ↓ | Component of the central pair apparatus of axonemes; function and structure of motile cilia; spermatogenesis; formation of sperm head and flagellum | III | |
| TCTE1* | Q5JU00 | 0.199 | ↓ | Nexin–dynein regulatory complex component; key regulator of ciliary/flagellar motility; microtubule sliding in motile axonemes; sperm motility | II | |
| CFAP43* | Q8NDM7 | 0.208 | ↓ | Sperm flagellum axoneme organization and function | III | |
| AK7* | Q96M32 | 0.258 | Ciliary structure and function | III | ||
| APAF1* | O14727 | 3.578 | Apoptosis trigger | IV | ||
| CATIP* | Q7Z7H3 | 0.293 | Ciliogenesis | IV | ||
| CATSPER2* | Q96P56 | 0.186 | Voltage-gated channel essential for sperm hyperactivation, acrosome reaction, and chemotaxis toward the oocyte | III | ||
| CFAP44* | Q96MT7 | 0.224 | Sperm flagellum axoneme organization and function | III | ||
| CFAP52* | Q8N1V2 | 0.283 | Ciliary and flagellar beating | III | ||
| CFAP58* | Q5T655 | 0.291 | Assembly and organization of the sperm flagellar axoneme | III | ||
| CFAP69* | A5D8W1 | 0.215 | Sperm flagellum assembly and stability | IV | ||
| CFAP70* | Q5T0N1 | 0.232 | Axoneme-binding protein; regulation of ciliary motility and cilium length | III | ||
| DNAH10* | Q8IVF4 | 0.268 | ATPase activity; sperm motility; sperm flagellar assembly | III | ||
| DNAH17* | Q9UFH2 | 0.300 | ATPase activity; outer dynein arms (…) in the sperm flagellum; sperm motility; sperm flagellar assembly and beating | III | ||
| FSIP2* | Q5CZC0 | 0.241 | Spermatogenesis | II | ||
| SMCP* | P49901 | 0.167 | Sperm motility | I | ||
| SPEF2* | Q9C093 | 0.215 | Development of sperm axoneme and manchette; sperm head morphology; adapter for dynein-mediated protein transport in spermatogenesis | III | ||
| SUN5* | Q8TC36 | 0.258 | Anchoring sperm head to the tail; attachment of the coupling apparatus to the sperm nuclear envelope | I | ||
| TTC29* | Q8NA56 | 0.252 | Axonemal and/or peri-axonemal assembly and structure; sperm flagellum assembly and beating | II |
- Note: Listed are sperm proteins with previous mention in the context of male fertility impairment (NCBI Online Mendelian Inheritance in Men [OMIM]) that showed at least threefold deviating abundances in infertile men diagnosed with oligoasthenoteratozoospermia (OAT) (false discovery rate [FDR] ≤ 0.05, t-test). Arrows indicate the direction of abundance changes in infertile men diagnosed with oligozoospermia (O) or oligoasthenozoospermia (OA). Changes refer to abundances in normozoospermic spermatozoa of fertile men. WDR66 corresponds to CFAP251. Asterisks highlight proteins included in the largest connected component shown in Figure 6. Categories on the left refer to transcript abundances reported in The Human Protein Atlas v22 (THPA): male germline-specific (I), male germline-biased (II), elevated in male germline and somatic cell types, which unlikely (III) or possibly were contained in the samples (IV).
| Symbol | Evidence | Panel |
|---|---|---|
| ACTL9 | Variants: fertilization failure, male infertility39 | Yes |
| CCIN |
Mutations: teratozoospermia, male infertility40 Sperm head-shaping factor essential for male fertility41 |
|
| CFAP47 | Variants: asthenoteratozoospermia, male infertility42 | Yes |
| CFAP65 |
Mutation: MMAF, male infertility43 Mutations: MMAF, male infertility44 |
|
|
WDR66 CFAP251 |
Deletion: MMAF, male infertility45 Loss: immotile spermatozoa, lacking mitochondria, male infertility46 |
Yes |
| DNAH1 | Mutations: MMAF47 | |
| SPEM1 |
Predictive value for sperm retrieval in azoospermia48 Lack in mice: aberrant cytoplasm rem., sperm deformation, male infertility49 |
|
| ACTL7A |
Disruption: acrosomal defects, early embryonic arrest50 Variants: total fertilization failure, male infertility51 |
|
| MNS1 |
Mutations: laterality defects, likely male infertility52 Variant: laterality defects, male infertility53 |
|
| PLCZ1 | Mutation: infertility, oocyte activation deficiency54 | Yes |
| PMFBP1 |
Mutations: ASS55 |
Yes |
| QRICH2 |
Mutations: MMAF58 Mutations: MMAF, male infertility59 |
Yes |
| SPAG17 | Mutation: asthenozoospermia60 | |
| TCTE1 | Variants: asthenozoospermia, male infertility61 | |
| CFAP43 |
Mutations: MMAF, male infertility62 Mutations: MMAF63 |
Yes |
| AK7 | Mutation: MMAF, male infertility64 | Yes |
| APAF1 | Raised testicular expression: male subfertility65 | |
| CATIP | Mutation: oligoasthenoteratozoospermia66 | Yes |
| CATSPER2 |
Non-syndromic male infertility67 Copy-number variant, reduced expression: idiopathic male infertility68 |
|
| CFAP44 |
Mutations: MMAF, male infertility62 Reduced expression: potential effect on sperm motility and morphology69 |
|
| CFAP52 | Exon 2 deletion: male infertility70, 71 | |
| CFAP58 | Variants: asthenozoospermia, flagellar axoneme + mitoch. sheath defects72 | Yes |
| CFAP69 | Absence: MMAF, male infertility73 | Yes |
| CFAP70 | Mutations: asthenoteratozoospermia, male infertility74 | Yes |
| DNAH10 |
Mutations: asthenoteratozoospermia, male infertility75 Variants: asthenoteratozoospermia, male infertility76 |
Yes |
| DNAH17 |
Mutations: asthenozoospermia, male infertility77 Variant: asthenozoospermia, flagella destabilization78 Intronic deletion: perturbed splicing, defect sperm flagella, male infertility79 |
Yes |
| FSIP2 | Mutations: MMAF80, 81 | Yes |
| SMCP |
Diff. expression in non-obstructive azoospermia versus oligoasthenozoospermia82 Deletion in mice: asthenozoospermia83 |
|
| SPEF2 |
Mutations: defects between flagella and cilia bridge, link of MMAF and PCD84 In mice: germ cell differentiation85 |
Yes |
| SUN5 | Mutations: ASS86, 87 | Yes |
| TTC29 | Mutations: asthenozoospermia, male infertility88 | Yes |
- Note: Evidence refers to men if not stated otherwise. “Yes” indicates inclusion in a male fertility panel of 81 genes (ID192.04).
- Abbreviations: ASS, acephalic spermatozoa syndrome; Diff., differential; mitoch., mitochondrial; MMAF, multiple morphological abnormalities of the flagella; PCD, primary ciliary dyskinesia; rem., removal.
| Symbol | UniProt ID | OAT | O | OA | Functional annotation | THPA |
|---|---|---|---|---|---|---|
| C2orf16 | Q68DN1 | 0.032 | ↓ | ↓ | High-abundance sperm protein in normozoospermic samples that achieved pregnancy20 | II |
| CYLC1* | P35663 | 0.056 | ↓ | ↓ | Architectural role during spermatogenesis; spermatid differentiation (UniProt) | I |
| SPATA31E1* | Q6ZUB1 | 0.061 | ↓ | ↓ | Spermatogenesis (UniProt) | II |
| SPATA31D1 | Q6ZQQ2 | 0.090 | ↓ | ↓ | Spermatogenesis (UniProt) | II |
| SPATA48* | A4D263 | 0.099 | ↓ | ↓ | Essential for normal spermatogenesis (UniProt) | II |
| EFHB | Q8N7U6 | 0.118 | ↓ | ↓ | Cilia- and flagella-associated protein 21 (HGNC) | II |
| FAM161A* | Q3B820 | 0.124 | ↓ | ↓ | Ciliogenesis (UniProt) | III |
| CYLC2* | Q14093 | 0.086 | ↓ | Architectural role during spermatogenesis; spermatid differentiation (UniProt) | I | |
| CCDC173 | Q0VFZ6 | 0.087 | ↓ | Cilia- and flagella-associated protein 210 (UniProt) | II | |
| SPEM3 | A0A1B0GUW6 | 0.092 | ↓ | Abundance varies with fertility differences in boars89 | I | |
| CFAP126* | Q5VTH2 | 0.098 | ↓ | Cilium basal body docking and positioning (UniProt) | II | |
| FAM71E2 | Q8N5Q1 | 0.105 | ↓ | Epigenetic transcriptional repression (UniProt) | II | |
| C1orf194* | Q5T5A4 | 0.105 | ↓ | Downregulated abundance in spermatozoa of patients with testicular cancer seminoma90 | II | |
| C1orf158* | Q8N1D5 | 0.110 | ↓ | Upregulated testicular expression in non-obstructive azoospermia91 | II | |
| DCDC2C | A8MYV0 | 0.120 | ↓ | Microtubule organization; sperm flagellum (QuickGO) | II | |
| EQTN* | Q9NQ60 | 0.123 | ↓ | Acrosomal membrane-anchored protein involved in (…) fertilization and in acrosome biogenesis (UniProt) | I | |
| SYPL1 | Q16563 | 0.107 | Downregulated testicular expression in non-obstructive azoospermia92 | IV | ||
| FAM205C | A6NFA0 | 0.118 | Abundance varies with fertility differences in boars89 | II |
- Note: Listed are sperm proteins without previous reference to male fertility impairment (NCBI Online Mendelian Inheritance in Men [OMIM]) that showed at least eightfold decreased abundances in oligoasthenoteratozoospermia (OAT) (false discovery rate [FDR] ≤ 0.05, t-test). Arrows indicate the direction abundance changes in spermatozoa of infertile men diagnosed with oligozoospermia (O) and oligoasthenozoospermia (OA). Changes refer to protein abundances in normozoospermic sperm proteins of fertile men. Asterisks highlight proteins included in the largest connected component shown in Figure 6. Categories on the left refer to transcript abundances reported in The Human Protein Atlas v22 (THPA): male germline-specific (I), male germline-biased (II), elevated in male germline and somatic cell types, which unlikely (III) or possibly were contained in the samples (IV).
| Symbol | Samples | Evidence | Direction |
|---|---|---|---|
| C2orf16 | Spermatozoa: 13 teratozoospermia versus 8 normozoospermia | Microarray | ↓93 |
| 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| 178 x testis: meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| CYLC1 | Spermatozoa: 13 teratozoospermia versus 8 normozoospermia | Microarray | ↓93 |
| 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| 178 x testis: meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| SPATA31E1 | 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 |
| 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| SPATA31D1 | Spermatozoa: 5 less versus 5 more fertilization-competent§ | PCR | ↓95 |
| SPATA48 | Testis: 6 azoospermia versus 6 normal fertile | PCR | ↓96 |
| EFHB | Spermatozoa: 13 teratozoospermia versus 8 normozoospermia | Microarray | ↓93 |
| 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| 178 x testis: pre-meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| FAM161A | 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 |
| CYLC2 | Spermatozoa: 13 teratozoospermia versus 8 normozoospermia | Microarray | ↓93 |
| 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| 178 x testis: meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| CCDC173 | 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 |
| SPEM3 | Spermatozoa: 4 less versus 4 more fertilization-competent+ | Mass spectrometry | ↓89 |
| CFAP126 | 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 |
| 178 x testis: meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| FAM71E2 | 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 |
| 178 x testis: meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| C1orf194 | Spermatozoa: 15 testicular cancer seminoma versus 15 control | Mass spectrometry | ↓90 |
| Spermatozoa: 20 asthenozoospermia versus 20 normozoospermia | Microarray | ↓9 | |
| C1orf158 | 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 |
| 178 x testis: meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| Spermatozoa: 13 teratozoospermia versus 8 normozoospermia | Microarray | ↓93 | |
| DCDC2C | Spermatozoa: 1 low-fertile male versus 1 high-fertile male+ | Microarray | ↓97 |
| EQTN | 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 |
| 178 x testis: meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| SYPL1 | Spermatozoa: 13 teratozoospermia versus 8 normozoospermia | Microarray | ↓93 |
| 178 x testis: pre-meiotic arrest versus three other diagnoses | Microarray | ↓94 | |
| FAM205C | 178 x testis: post-meiotic arrest versus three other diagnoses | Microarray | ↓94 |
| 178 x testis: meiotic arrest versus three other diagnoses | Microarray | ↓94 |
- Note: Comparisons mostly refer to men and exceptionally to domestic pig (+) and horse (§).
- Abbreviation: PCR, polymerase chain reaction.
3.3 Quantitative MS analysis
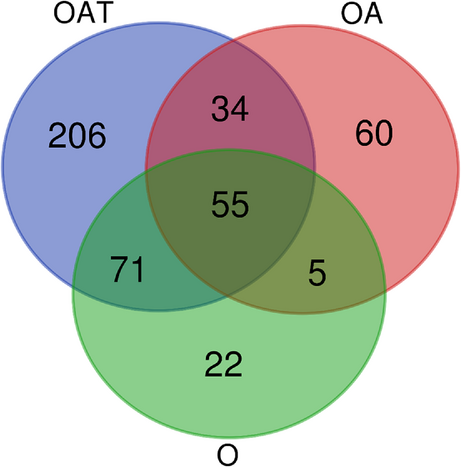
The pipeline applied unveiled numerous protein groups with at least threefold differential, mostly lowered, abundances (FDR ≤ 0.05, each; t-test) in spermatozoa of infertile men diagnosed with O, OA, and OAT, relative to normozoospermic spermatozoa of normal-fertile men (Table S4). Thus, 133 of 153 (87%) differentially abundant protein groups exhibited downregulation in O spermatozoa (O list). With 154, the total number of protein groups varying in abundance was almost the same in the OA versus normozoospermia comparison (OA list), whereby most of these showed declines in abundance under infertility once more (N = 110; 71%). The OAT versus normozoospermia comparison (OAT list) revealed the highest number of differentially abundant protein groups (N = 368), with lowered levels predominating in the infertility cohort again (N = 315; 86%) (Table S4). Fifty-five protein groups were shared among the three O-associated lists. In addition, there was an increasing number of unique protein groups with growing complexity of the diagnosis, and thus from the O via the OA to the OAT list (Figure 3). Also, the number of overrepresented GOs (FDR ≤ 0.001, each) was much higher in the OAT list (N = 72) than in O (N = 26) and OA (N = 20) lists. Nevertheless, cilium-dependent cell motility yielded the largest fraction of GOs in all three infertility diagnoses. The three lists also had in common that 75% of the overrepresentations pertained to the structure and function of cilia or the sperm flagellum. Additional overrepresentations referred to male gamete generation and fertilization (O, OA, and OAT lists), cAMP signaling (OA and OAT lists), and sodium–potassium exchange (OAT list) (Figure 4).
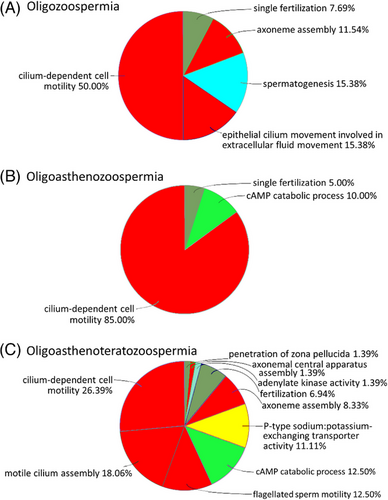
Twelve percent of the altogether 198 genes with OMIM entries relating to male fertility impairment had counterparts in the most comprehensive of our three lists, the OAT list (Table S5). Broken down to individual aberrations from normozoospermia, we observed the following pattern: the gene list compiled from OMIM entries on teratozoospermia matched to 10% (3 of 29) with genes or proteins in our OAT list. In OMIM lists for asthenozoospermia and teratozoospermia, 17% (12 of 70) and 35% of the genes (23 of 66) had matches in our OAT list. Thus, the proportion of matching genes in all three OMIM lists considerably exceeded the random expectation of <1%, as given by the total number of genes in single OMIM lists (N = 23–70) divided by 20,471 protein-coding human genes according to ENSEMBL Genes 106: GRCh38.p13.
3.4 Candidate markers of male fertility
Thirty-one sperm proteins in our OAT list had entries in the OMIM database suggestive of an involvement in male fertility impairment (Table 1). Their GOs related to sperm flagellum assembly, structure and beating, as well as male gametogenesis. We refer to these proteins as to validated candidate markers in the following. Out of these, apoptosis trigger APAF1 was the only one showing higher abundance in OAT than normozoospermic spermatozoa (Table 1). The remaining proteins with previous mention relating to male fertility impairment exhibited reduced levels under OAT. Seven of these displayed decreased abundances across all three O-associated diagnoses: actin-like protein 9 (ACTL9), calicin (CCIN), CFAP47, CFAP65, CFAP251 (WDR66), dynein axonemal heavy chain 1 (DNAH1), and sperm maturation protein 1 (SPEM1) (Table 1). In-depth search for previous evidence (Table 2) revealed that most validated candidate markers were previously associated with malformed and dysfunctional sperm flagella and impaired sperm motility. Corresponding terms included multiple morphological abnormalities of the flagella (MMAF) and asthenozoospermia (N = 20). This was followed by associations with morphological defects such as acephalic spermatozoa syndrome (ASS), acrosomal defects, and lacking mitochondria (N = 10). The least connections were found to reduced sperm count (N = 3). Notably, 18 of the validated candidate markers of male fertility were covered by an 81 gene panel and hence are already in use in male fertility diagnostics (Table 2).
Lack of OMIM entries relating to male fertility impairment let us refer to a group of 18 sperm proteins with extremely deviating abundances under infertility as to novel candidate markers. These exhibited at least eightfold reduced abundances in the OAT–normozoospermia comparison (Table 3). According to the data available, the novel candidate markers engage in spermiogenesis, including ciliogenesis and acrosome biogenesis. For all novel candidate markers, we found previous indications of downregulated expression under reduced male fertility. For example, no SPATA48 transcript was determined in testicular samples of six azoospermic men, while corresponding transcripts were highly abundant in healthy subjects (Table 4). Furthermore, seven proteins had significantly reduced abundances in O and OA spermatozoa, and, hence, might elucidate the deeper causes of male fertility impairment whenever reduced sperm count is suspected to play a role.
There were three protein groups, each containing two proteins, with decreased abundances in spermatozoa under male infertility (Table 5). Half of these proteins had OMIM entries relating to male fertility. Until clarification whether one or both group members occur at lowered abundances in spermatozoa of infertile men, the corresponding proteins lend themselves less readily as candidate markers.
| Symbol | ID | OAT | O | OA | Functional annotation (UniProt) | THPA |
|---|---|---|---|---|---|---|
|
SPATA31D3, SPATA31D4* |
P0C874, Q6ZUB0 |
0.091 | ↓ | ↓ | Spermatogenesis |
I I |
|
CFAP45*, SNHG28 |
Q9UL16, P0DPA3 |
0.101 | Dynein ATPase-dependent ciliary and flagellar beating |
III No entry |
||
|
DNAH8*, DNAH14 |
Q96JB1, A0A804HLD3 |
0.318 | ATPase activity; outer dynein arms (…) sperm flagellum; sperm flagellar assembly; sperm motility |
II III |
- Note: The list contains protein groups showing at least three- or eightfold differential abundances in spermatozoa of infertile men diagnosed with oligoasthenoteratozoospermia (OAT) (false discovery rate [FDR] ≤ 0.05, t-test). Arrows indicate the direction of abundance changes in spermatozoa of infertile men diagnosed with oligozoospermia (O) and oligoasthenozoospermia (OA). Changes refer to abundances in normozoospermic spermatozoa of fertile men. Commas separate members of single protein groups. Asterisks highlight proteins included in the largest connected component shown in Figure 6. Categories on the left refer to transcript abundances reported in The Human Protein Atlas v22 (THPA): male germline-specific (I), male germline-biased (II), and elevated in male germline and somatic cell types, which unlikely were contained in the samples (III).
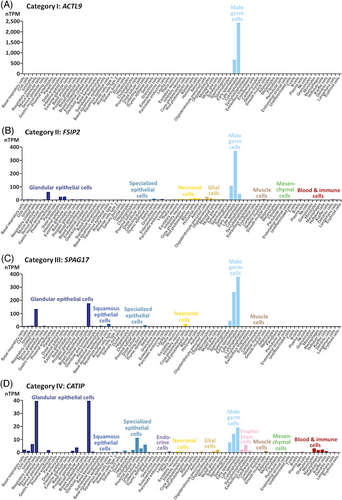
Downstream, we checked for potential somatic expression of the genes to the proteins in Tables 1, 3, and 5. According to version 22 of The Human Protein Atlas, transcripts of most of the genes with validated male fertility relevance (Table 1) were specific to (category I: seven genes) or much more frequent in male germline (category II: six genes), or transcript abundance was elevated in male germline and somatic cell types, which will not have contaminated our samples (category III: 15 genes). The latter was exemplified in respiratory ciliated and endometrial ciliated cells (Figures 5 and S1). Genes of only few validated candidate markers showed the expected expression in male germline and additionally displayed elevated transcript abundances in somatic cell types that could have been contained in the samples analyzed (category IV: three genes). Prevalence of categories I–III was reproduced in genes encoding novel candidate markers (Table 3 and Figure S2). Most of the coding genes displayed exclusive (category I: four genes) or skewed expression in male germline (category II: 12 genes), and one gene additionally exhibited raised transcript amounts in somatic cells, which unlikely were contained in the samples (category III: one gene). The transcriptional profile of only one of the novel candidate markers allowed for the possibility that somatic cell types could have been included in the sample in addition to the targeted spermatozoa (category IV). The genes underlying protein groups even spread without exception across categories I–III (Table 5 and Figure S3). Thus, for only few of the proteins contamination with somatic cells might have contributed to the finding of differential abundance. But also, these few ones had OMIM entries indicating male fertility relevance (Table 1: APAF1, CATIP, CFAP69) or were already found to be downregulated in spermatozoa under male infertility (Table 3: SYPL1). We therefore assume that contaminations of our sperm samples with somatic cells should not have compromised present results.
3.5 The male infertility network
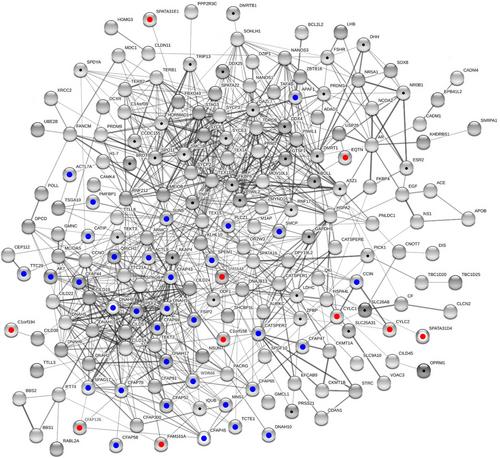
For all protein samples considered, STRING gathered more PPIs than to be expected for the proteins mapped (FDR < 0.001, each). In addition, all network parameters rose with growing number of proteins included (Tables 6 and S6). Thus, STRING built a network of 50 nodes, thereof 66% contained in the largest connected component (LCC), and 75 edges when run on the proteins in Tables 1, 3, and 5. Higher coherence was reached upon addition of 41 candidate markers for which we had previously calculated elevated fertility relevance probability.19 Especially, the number of disconnected nodes decreased, despite an almost doubled number of proteins mapped (N = 91). With 208, this network had a disproportionally larger number of edges. Correspondingly, the number of PPIs per node was higher in the expanded network (node degree = 4.57) than in the one restricted to the proteins highlighted in the present study (node degree = 3.00). Addition of all other proteins with OMIM entries suggestive of male fertility relevance more than doubled the number of nodes to 225. The total number of PPIs increased to 891, and the average node degree to 7.92 (Table 6). The corresponding LCC was comprised of 192 nodes (Figure 6), thereunder half of the novel candidate markers and all of the validated candidate markers. Also included in the LCC were 37 out of the abovementioned 41 proteins with increased fertility relevance probability,19 and most of the proteins with OMIM entries pointing to male fertility relevance. We refer to this LCC as to the male infertility network.
| Dataset source | NNodes | NEdges (Nexp) | NNodes in LCC | %Nodes in LCC | Average nd | EET (FDR) |
|---|---|---|---|---|---|---|
| Present study | 50 | 75 (2) | 33 | 66 | 3.00 | <0.001 |
| Present study + FRP | 91 | 208 (13) | 75 | 82 | 4.57 | <0.001 |
| Present study + FRP + OMIM | 225 | 891 (132) | 192 | 85 | 7.92 | <0.001 |
- Note: Networks were reconstructed from the proteins carved out in the present study (Tables 1, 3, 5), previous inference of fertility relevance probability (FRP)19, and database screening (NCBI Online Mendelian Inheritance in Men [OMIM]). Numbers of nodes refer to the proteins mapped by STRING v11.5.
- Abbreviations: EET, edge enrichment test; FDR, false discovery rate; LCC, largest connected component; Nexp, expected number of edges; nd, node degree.
4 DISCUSSION
4.1 The human sperm proteome and its coding genes
According to present MS analysis, about 6981 genes participate in the human sperm proteome (Tables S1 and S2). This represents a considerable increase compared with previous estimates from MS studies, which had determined 344421 and 471822 sperm proteins. The current number also exceeds the 6871 sperm proteins as gathered in a recent survey.8 Merging the compilation from the latter study with present MS results, about 9300 genes might participate in the human sperm proteome. This would be ca. 45% of the approximately 20,500 coding human genes according to genome annotation ENSEMBL Gene 106: GRCh38.p13. The complexity of the sperm proteome will be higher, considering diversification by alternative splicing and post-translational modification.29, 98, 99
Overall shorter cDNAs, as observed in this study, could reflect adaptation to better swimming performance. In line with such possibility, there is previous evidence that selection for increased motility might drive changes in sperm morphology.100, 101 This might involve remodeling of protamines which are well-known for evolving under sexual selection.102 In the present reconstruction of the human sperm proteome, importance of sperm motility was reflected in implications of numerous coding genes in cilium assembly and function (Figure 2). GOs referring to cellular responsiveness and adhesion were also conclusive considering the challenges spermatozoa have to cope with.103, 104 This might also be true for annotations referencing the immune system. In fact, male fertility and fertilization require evasion from the own immune response and the one of the partner.99, 105 Still, corresponding GO groups contained genes encoding proteins with clear fertility relevance, such as SPATA20,11, 106 ZPBP,107 and SPAG4108 (Table S3). Thus, reference to the immune system should not be overestimated in our opinion. Not least, present GO analyses (Figure 2) confirmed that spermatozoa contain proteins that are known primarily for their roles in male gametogenesis.109
4.2 Molecular causes of male fertility impairment
Results of present overrepresentation tests revealed that the fewer spermatozoa produced under O, OA, and OAT carry signatures of disturbed gametogenesis. Functional annotations overrepresented in differentially abundant proteins were further consistent with impairment of sperm motility and fertilization competence in infertile men (Figure 4). This includes proteins involved in cAMP signaling and sodium–potassium exchange. The first is crucial for all steps of sperm function in mammals,110 and importance for motility and fertilization has been assigned to the latter.111Significance of proper sperm motility for reproduction112 was additionally reflected in frequent references to the structure and function of the cilium or flagellum (Figure 4). The pattern was even more strongly reflected in the data on the individual proteins and coding genes. Thus, for the validated and novel candidate markers, there was evidence of involvements in malformations such as ASS and MMAF and, related to the latter, impaired motility or asthenozoospermia (Tables 1–4). This suggests that reduced sperm count could be less decisive in O and syndromes including it. The ultimate reason might rather be that the fewer spermatozoa produced are dysfunctional to an increased proportion. This seems particularly noteworthy given that the samples analyzed had undergone a swim-up, meaning that the spermatozoa represented a selection of spermatozoa with increased fertilization competence. Even these would be functionally compromised according to our results, to an increased proportion at least. This could also explain why lowered spermiogram parameters can reduce success rates in ARTs.113, 114
4.3 Toward a marker panel of male fertility assessment
Quantitative MS analysis of the entire sperm proteome might complement diagnostics of the male fertility status in the future. If it comes to this point, special attention may be paid to the abundances of particular sperm proteins. Estimating the fertility status from differential abundances of sperm proteins on scales as found here (N = 153–368) should then be more feasible than based on numbers in the range of 1000 or higher as previously reported.21, 22 This was achieved by applying a more conservative threshold of differential abundance in the present study (at least 3×) than was the case in both referenced investigations (at least 1.5×).21, 22 Still, the definition of a further reduced number of male fertility markers remains to be a worthwhile goal for standardized fertility testing.19, 28, 115 The proteins summarized in Tables 1 and 3, and to a lesser extent in Table 5, may contribute to this. It will be interesting to validate in future independent studies the corresponding proteins using alternative methods such as western blot.
Diagnostic potential should especially have the 31 sperm proteins with previous mention in the OMIM database hinting to roles in the etiology of male infertility (Table 1). Increased importance of these validated candidate markers for reproduction is underscored by transcript data. Indeed, we found expression of most of the coding genes to be tailored to male germline (Figure S1). Numerous associations with male fertility impairment lend additional support for their marker potential, as did their already-mentioned involvements in sperm flagellum structure and functioning, sperm head formation, and spermiogenesis (Tables 1 and 2). Against this background, it also seems plausible for most validated candidate markers that a decrease in abundance should be detrimental for male fertility maintenance. There were only two exceptions from the pattern. Exceptional in terms of function was 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1). The protein proposedly belongs to the sperm factors transferred to the zygote and as such is likely to initiate the development of the zygote into the blastocyst.116-118 Consistently, a PLCZ1 variant in an infertile man was found to associate with oocyte activation deficiency.54 Thus, infertility at PLCZ1 shortage could hint to disturbed development following fertilization of the ovum. Still, PLCZ1 seems to be also important for spermiogenesis considering spermatogenic defect in male Plcz1 knockout mice.119 The second exception from the above pattern refers to the direction of abundance change in reduced-relative to normal-fertile men. In fact, apoptotic protease-activating factor 1 (APAF1) was the only validated candidate marker showing increased abundance under infertility. Although exceptional in the present study, the present finding fits previous evidence of raised testicular abundance of APAF1 transcript in men with impaired versus normal spermiogenesis.65 It appears to be plausible for an apoptosis trigger120 that oversupply negatively affects sperm number and viability.20
Seven of the proteins, to which we here refer to as validated candidate markers, displayed deviant abundances under O, OA, and OAT (Table 1): ACTL9, CCIN, CFAP47, CFAP65, CFAP251 (WDR66), DNAH1, and SPEM1. Functional data would be in line with a predictive value of corresponding protein abundances whenever a reduction in sperm count is observed. Thus, SPEM1 should have importance for proper sperm formation including cytoplasm removal as suggested by deformed spermatozoa and infertility in male knockout mice.49 Furthermore, ACTL9 plays a significant role in the fusion of proacrosomal vesicles and the anchoring of the acrosome to the perinuclear theca during spermiogenesis. Correspondingly, there are variants that associate with fertilization disorders and male infertility.39 CCIN also localizes to the acrosomal area, thereby showing affinity to F-actin.121 As a basic cytoskeleton protein of the mammalian sperm head, absence or dysregulation of CCIN is known to cause sperm malformation.122 Relevance for proper flagellum formation and functioning is further inherent to three cilia and flagella-associated proteins (CFAP47, CFAP65, CFAP251/WDR66) and DNAH1.42, 43, 45, 46, 91, 123, 124 Nevertheless, all sperm proteins in Table 1 could be implicated in the development of male fertility disorders, and hence might be further evaluated for their potential as male fertility markers. In support of such utilization, the coding genes of 18 out of the 31 validated candidate markers are already included in an 81 gene panel (ID192.04) for male fertility testing (e.g., www.zhma.de).
Diagnostic potential might also have 18 proteins displaying at least eightfold and thus extreme deregulation of abundance (Table 3). We refer to these proteins as novel candidate markers because OMIM search did not uncover respective previous entries. Nevertheless, downregulation of testicular and spermatozoic expression under reduced male fertility was documented for the coding genes to all novel candidate markers (Table 4). Predictive power of corresponding protein levels would also be consistent with the functional and transcriptional data available, as will be detailed below in the ones showing reduced levels in all three diagnoses including reduced sperm count. In case of SPATA31E1, SPATA31D1, and SPATA48, relevance for male fertility is reflected by transcript enrichment in the male germline (Figure S2). Transcriptional data additionally suggest that EF-hand domain family member B (EFHB, alias CFAP21), chromosome 2 open reading frame 16 (C2orf16, alias CB016), and family with sequence similarity 161 member A (FAM161A) participate in male fertility maintenance (Figure S2). Indeed, EFHB is a regulator of store-operated Ca2+ entry (SOCE) impairment of which causes male infertility in the murine model.125, 126 Furthermore, FAM161A is a component of the cilia–basal body complex127 modulating mammalian sperm movement.127, 128 Another protein with strongly deregulated abundance in association with O, cylicin 1 (CYLC1), is involved in the formation of the sperm head.129 As in several other novel candidate markers, CYLC1 transcription is highly enriched in male germ line, suggestive of special importance for male fertility (Figure S2).
5 CONCLUSIONS
Present high-resolution mass spectrometry analyses of sperm samples from 38 reduced- and 38 normal-fertile men uncovered proteins of about 7000 genes (Table S2). The functional data available for differentially abundant proteins suggests that spermatozoa of infertile donors diagnosed with oligoasthenoteratozoospermia, oligoasthenozoospermia, and oligozoospermia carry proteomic signatures of disturbed gametogenesis. Our results further imply that the fewer spermatozoa in these diagnoses are dysfunctional to an increased proportion, with motility and fertilization ability being compromised. Thus, a reduction in sperm count might have been less decisive for the development of infertility in the corresponding subjects than the fact that the spermatozoa produced were dysfunctional to an increased proportion. While this was to be expected for both syndromes it was not for cases in which pure oligozoospermia was diagnosed. This sheds new light on the challenges in selecting proper spermatozoa for in vitro fertilization and intracytoplasmic sperm injection based on microscopic examination.113, 114
Quantitative analysis led to the identification of sperm proteins the abundances of which might have predictive value in fertility diagnostics. As detailed above, the respective proteins had been determined in considerably high numbers of peptides, suggestive of highly confident identifications. Marker potential should especially have the ones for which the coding genes already had entries in the Online Mendelian Inheritance in Men (OMIM) database relating to roles in male fertility impairment (Tables 1 and 2 and Figure S1). Such applied prospect is matched by the fact that the coding genes of about two-thirds of the proteins are included in a genotyping panel used in fertility testing (ID192.04). Yet proteins for which we observed extremely reduced levels under infertility (Table 3) might also be considered for their potential in fertility diagnostics. In support of such usability, expression of the coding genes was previously found to be reduced in testes and spermatozoa of men with impaired fertility (Table 4; see also Figure S2). Members of deregulated protein groups might be evaluated for utilization in fertility testing too (Table 5 and Figure S3).
Our results further illustrate that most of the proteins highlighted above are interconnected with other players in the development of male infertility. Indeed, we observed a growing connectivity with increasing number of prospective male fertility markers included in network reconstructions (Figure 6 and Table 6). We expect the male infertility network to grow with the addition of further (candidate) markers.9, 21, 130 For example, future network analyses may include male fertility markers in the seminal plasma proteome.9, 21, 131, 132 With the completion of the network, we should increasingly become able to comprehend the molecular mechanisms behind the development of male infertility. This may open new perspectives in the diagnosis and treatment of male infertility.133
AUTHOR CONTRIBUTIONS
Thomas Greither and Holger Herlyn conceived the study. Thomas Greither and Hermann M. Behre recruited and characterized the participants. Falk Butter, Mario Dejung, and Holger Herlyn performed the experiments. Holger Herlyn analyzed the data and drafted the manuscript. Thomas Greither, Hermann M. Behre, Falk Butter, and Holger Herlyn critically revised the manuscript. All authors approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank all participants for their contribution to this study. Constanze Kloß is thanked for excellent technical assistance. We gratefully acknowledge constructive comments in the review process the implementation of which greatly improved our study.
Open access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest to disclose.
FUNDING INFORMATION
This work received no external funding.