BK Virus Encoded MicroRNAs Are Present in Blood of Renal Transplant Recipients With BK Viral Nephropathy
Abstract
BK viral infection is an important cause of renal transplant dysfunction and failure. Current strategies utilize surveillance for infection with DNA polymerase chain reaction assays and modulation of immunosuppression. Many viruses including polyomaviruses encode microRNAs (miRNAs). We have detected BK virus (BKV) encoded miRNAs in the blood of infected renal transplant recipients, and see a strong correlation between BKV encoded miRNA and BKV DNA in blood and a relationship between levels of bkv-miR-B1-5p and the presence of biopsy-proven BK viral nephropathy. Further research is needed to determine whether the detection of this and other virally encoded miRNAs may be useful in the diagnosis of active viral replication.
Abbreviations
-
- BKV
-
- BK virus
-
- BKVN
-
- BK viral nephropathy
-
- cDNA
-
- complementary DNA
-
- Cq
-
- quantification cycle
-
- EBV
-
- Epstein–Barr virus
-
- eGFR
-
- estimated GFR
-
- JCV
-
- JC virus
-
- miRNA
-
- microRNA
-
- PCR
-
- polymerase chain reaction
-
- qRT-PCR
-
- quantitative real-time reverse transcriptase polymerase chain reaction
-
- ROC
-
- receiver operating characteristic
-
- RT-PCR
-
- reverse transcriptase polymerase chain reaction
-
- sMDRD
-
- simplified Modification of Diet in Renal Disease
-
- TAg
-
- viral protein T antigen
Introduction
Two polyomaviruses, BK virus (BKV) and JC virus (JCV), establish a latent infection in 65–90% of humans 1, 2. BKV is the causative agent of BK viral nephropathy (BKVN) in kidney transplant recipients 3. BKVN can lead to deterioration in allograft function and graft failure 4, 5. BKV reactivation occurs commonly after renal transplantation and is identified in the urine of 30–50% of patients at 3 months posttransplantation 6. Viral DNA in blood and urine is usually detected by quantitative polymerase chain reaction (PCR) 7. Progression from detection of viral DNA in urine to blood occurs in approximately 15% of kidney transplant recipients and is a harbinger of BKVN and deterioration of renal function 5, 7, 8.
MicroRNAs (miRNAs) are noncoding RNAs of 20–22 nucleotides. miRNAs regulate the translation and stability of mRNA by binding to complementary “target” mRNA sequences. Lost or enhanced expression of miRNAs is associated with fundamental cellular processes 9. Members of several virus families have been reported to encode miRNAs 10. The functions of the majority of viral miRNAs are unknown, though some viral miRNAs regulate host gene expression, viral gene expression 11 and transition from latency to lytic replication and can attenuate immune responses 12-14. Virally encoded miRNA expression has been described in viral replication. miRNA encoded by the Merkel cell polyomavirus has been detected in specimens of Merkel cell carcinoma 15, and miRNAs encoded by Epstein–Barr virus (EBV) have been detected in EBV-associated nasopharyngeal carcinoma 16.
BKV encoded miRNAs have been demonstrated to target the viral protein T antigen (TAg) with perfect complementarity to the 3p coding end of the TAg mRNA 11, 17. One of the major functions of TAg is driving DNA replication of the viral genome at the origin of replication 18. Furthermore, BKV miRNA can target stress-induced protein ULBP3, which is recognized by the natural killer cells receptor NKG2D 13 to potentially avoid NKG2D-mediated elimination.
It is not known whether infected renal cells release BKV miRNA (bkv-miR-B1) or whether this miRNA can be detected in the blood or urine of infected patients. In this study, we report the novel observation that BKV miRNA is expressed during replication and can be detected in blood. We also find a correlation between BKV miRNA and BKV DNA in blood and increased levels in patients with biopsy-proven BKVN.
Materials and Methods
Study population
The files of the Renal Transplant Patients in South Australia registry were reviewed to identify adult patients (age > 18 years) with BKV DNA in the blood (ascertained by BK DNA PCR) following a kidney transplant between January 2008 and May 2013. Thirty-one patients with BKV DNA in blood and stored blood samples were studied. Five patients with previous blood BK DNA positive but now negative were included for comparison. Ten patients who never had detectable BKV in the blood and urine from transplant until May 2013 were selected as a control group.
Patients' estimated GFR (eGFR) was calculated using the simplified Modification of Diet in Renal Disease (sMDRD) formula. The diagnosis of BKVN was made by the presence of viral cytopathic effect in renal tubular cells with interstitial mononuclear inflammatory cell infiltrates and/or the presence of homogenous intranuclear inclusion bodies. This was supplemented by immunohistochemistry with Simian virus 40 staining 19.
This study was approved by both the Southern and Central Adelaide Local Health Network Human Research Ethics Committees (Approval No: 326.11 and 110918, respectively).
Study method
Design of primers and PCR for BKV and JCV DNA
The full method has been described previously 20. Primers and probes for BKV were designed to target the VP2 region. Total RNA was extracted from 0.25 mL of thawed plasma using Trizol LS (Invitrogen, Life Technologies, Carlsbad, CA). The RNA pellet was resuspended in 20 μL of RNase-free water and stored at −80°C.
Quantitative real-time reverse transcriptase PCR
BKV miRNA expression was assessed by quantitative real-time reverse transcriptase PCR (qRT-PCR) using human TaqMan miRNA assays (Applied Biosystems, Foster City, CA). The reverse transcriptase reaction using 4 µL of RNA sample was performed with TaqMan miRNA Reverse Transcription Kit and TaqMan miRNA-specific primers (assay IDs: hsa-miR-16:000391, bkv-miR-B1-5p:007796, bkv-miR-B1-3p:006801; Applied Biosystems). PCR reactions were pipetted in triplicate using a Qiagen QIAgility robot (v4.15.1; Melbourne, Australia). The qRT-PCR reaction contained 1 μL of reverse transcription product, 1× TaqMan Universal PCR mastermix, No AmpErase UNG and 0.5 μL of primer mix. The complementary DNA (cDNA) was amplified using a Rotor-Gene Q Thermocycler (Qiagen). Reactions were incubated at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 60 s. Quantification cycle (Cq) values were calculated using the Rotor-Gene Q software (v2.0.2).
A control plasma sample was spiked with a known concentration of synthetic BKV miRNA mimic duplex oligonucleotides (bkv-miR-B1; Shanghai GenePharma, Shanghai, China). The sequence of the duplex was 5′-AUCUGAGACUUGGGAAGAGCAU and 5′-UGCUUGAUCCAUGUCCAGAGUC. cDNA was synthesized from 4 µL of total RNA sample using a specific primer for bkv-miR-B1-5p.
Statistical analysis
A one-way analysis of variance was used to compare the means of the patient characteristics (Table 1). The Spearman's Rho nonparametric correlation coefficient was used to assess correlations between BKV miRNA expression, BKV DNA viral load and laboratory measurements. The Kruskal–Wallis nonparametric test was used to compare BKV miRNA expression across all five clinical groups. A Mann–Whitney nonparametric test was used to assess the significance of BKV miRNA expression in pairwise comparisons of clinical groups when p < 0.05 across all five groups for the Kruskal–Wallis test. For all statistical tests, two-sided p < 0.05 was considered statistically significant.
| Clinical category | Biopsy-proven BKVAN | No BKVAN on biopsy | BK PCR DNA positive/not biopsied | Previous BK PCR DNA positive now negative | Negative BK DNA PCR | p-Value |
|---|---|---|---|---|---|---|
| BKVAN | No BKVAN | BK positive/no biopsy | Previous BK positive | BK negative | ||
| BK PCR DNA detected | + | + | + | − | − | – |
| Renal biopsy undertaken | + | + | − | − | − | – |
| Renal biopsy showing BKVAN | + | − | − | − | − | – |
| Number of patients | 7 | 13 | 11 | 5 | 10 | – |
| Age (years) | 63 ± 8 | 50 ± 16 | 57 ± 11 | 60 ± 8 | 50 ± 12 | 0.13* |
| Sex (male:female) | 2:5 | 8:5 | 9:2 | 4:1 | 5:5 | – |
| Time since transplant (median, months) | 18 ± 43 | 5 ± 4 | 4 ± 4 | 11 ± 17 | 2 ± 10 | 0.003* |
| Serum creatinine1 (µmol/L) | 175 ± 132 | 168 ± 79 | 124 ± 58 | 148 ± 43 | 130 ± 75 | 0.57* |
| eGFR1 (mL/min/1.73 m2) | 50 ± 39 | 42 ± 18 | 61 ± 24 | 43 ± 10 | 56 ± 24 | 0.36* |
| Serum creatinine2 (µmol/L) | 155 ± 103 | 140 ± 56 | 121 ± 56 | 140 ± 50 | 115 ± 43 | 0.67* |
| eGFR2 (mL/min/1.73 m2) | 39 ± 21 | 48 ± 14 | 51 ± 15 | 47 ± 15 | 53 ± 11 | 0.47* |
| Hemoglobin1 (g/L) | 111 ± 15 | 113 ± 19 | 128 ± 19 | 131 ± 10 | 112 ± 14 | 0.039* |
| Platelet count1 (×109/L) | 235 ± 40 | 208 ± 56 | 209 ± 55 | 185 ± 30 | 259 ± 67 | 0.083* |
| BK copy number PCR1 (/mL) | 2 666 857 ± 1 859 790 | 406 445 ± 765 001 | 157 973 ± 240 871 | 0 | 0 | <0.001* |
- BKVN, BK viral nephropathy; eGFR, estimated GFR; PCR, polymerase chain reaction.
- Gender, age, creatinine, eGFR (sMDRD; simplified Modification of Diet in Renal Disease formula), hemoglobin, platelet count and BK PCR DNA viral load of the 46 patients are shown. Values are means ± SD.
- 1 At time of sample acquisition for determination of BK PCR DNA and BK miRNA.
- 2 One-year after sample acquisition.
- * p-Value generated from one-way analysis of variance; significant p-values (<0.05) shown in bold.
We assessed the correlation of plasma BKV miRNA, BKV DNA viral load, and the detection of BKVN. Each marker was assessed separately using logistic regression and the C-statistic (area-under receiver operating characteristic [ROC] curve). Tests of equality of the C-statistic between measures were performed using the algorithm suggested by DeLong et al 21. We used plots of the predicted probabilities of the logistic regression to determine the optimal cut-point of each measure in providing both high sensitivity and high specificity and calculated sensitivity and specificity based on these cut-points.
Results
The detection of BKV miRNA
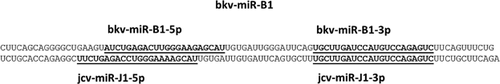
BKV encodes one precursor miRNA, bkv-miR-B1, which generates two mature miRNAs, 3p and 5p. The 3p miRNA is conserved between JCV and BKV. However, the sequence of the mature 5p miRNA generated by BKV and JCV differs, providing a mean of differentiating between these two viral infections (Figure 1). A nucleotide blast search (Basic Local Alignment Search Tool; http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) using a 102-nucleotide query corresponding to the precursor stem-loop sequence bkv-miR-B1 revealed perfect sequence conservation in all (over 100) complete BKV genome sequences in GenBank (www.ncbi.nlm.nih.gov/genbank). To examine for bkv-miR-B1, we used qRT-PCR using human TaqMan microRNA assays (Applied Biosystems). As a positive control and to enable approximate quantification of oligonucleotide copy number, plasma was spiked with a known concentration of synthetic BKV miRNA oligonucleotides (bkv-miR-B1) or with a similar JCV oligonucleotide. Initial assays confirmed the ability of the TaqMan reverse transcriptase PCR (RT-PCR) assays to detect both bkv-miR-B1-5p and bkv-miR-B1-3p. In keeping with the identity of the 3p miRNAs, expression of the JCV encoding oligonucleotide could also be detected by the bkv-miR-B1-3p RT-PCR assay but the bkv-miR-B1-5p assay was 500-fold more sensitive (data not shown) in detecting the BK miRNA transcript than the JC encoded transcript.
The detection of BKV miRNAs in blood from patients with BK replication
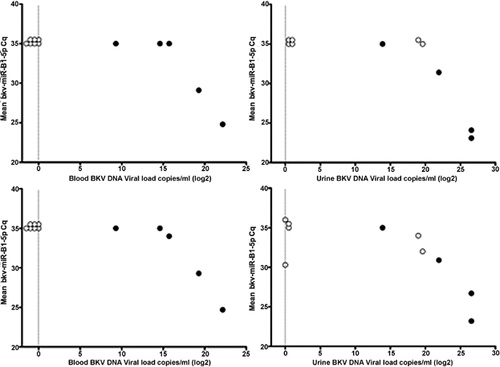
The presence of BK miRNA (bkv-miR-B1-3p and bkv-miR-B1-5p) was examined by TaqMan qRT-PCR in RNA purified from blood and urine of renal transplant recipients with BK DNA detected by PCR and in controls (renal transplant recipients with no detectable BK DNA). We were able to detect bkv-miR-B1-3p and bkv-miR-B1-5p in both blood and urine of some, but not all, patients with detectable BK DNA (Figure 2) with Cq values ranging from 24 to 34. A significant negative correlation was found between the qRT-PCR Cq value and the detection of BKV DNA in blood and urine suggesting a higher level of bkv-miR-B1-5p and bkv-miR-B1-3p in patients with higher BKV DNA levels (Figure 2). Cq values are inversely proportional to the amount of target nucleic acid in the sample, that is, the lower the Cq, the greater the amount of BKV miRNA in the sample. The bkv-miR-B1-3p miRNA was detected in the urine of two control patients with positive urine BKV DNA but negative blood BKV DNA. We were unable to detect BK miRNA in control patients without BKV infection other than amplification of bkv-miR-B1-3p with a Cq of 30.3 in the urine from a single control patient with negative BKV DNA (Figure 2).
Optimization of bkv-miR-B1-5p qRT-PCR assay
Given the ability of the bkv-miR-B1-5p qRT-PCR assay to detect BKV miRNA but not the JCV encoded 5p miRNA, we focused on this assay. Stem-loop primed TaqMan assays for miRNA detection are specific for mature forms of bkv-miR-B1-5p, and in keeping with design of the assay to detect RNA, positive samples assayed without the reverse transcriptase did not show bkv-miR-B1-5p PCR amplification, in accord with reports of inability to amplify genomic DNA sequences using this method 22.
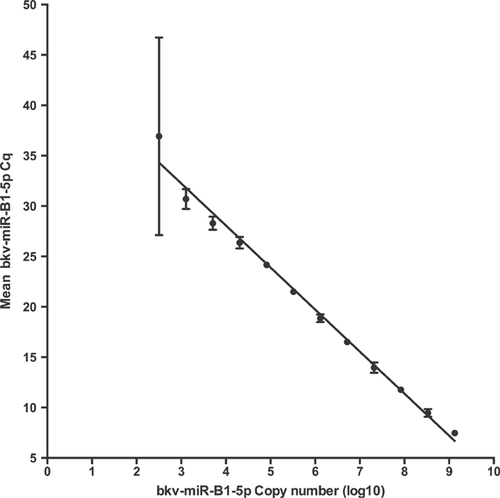
To provide an approximate quantification of the number of molecules of bkv-miR-B1-5p, we undertook a titration series of synthetic oligonucleotide duplex BKV miRNA mimic. A control plasma sample was spiked with a known concentration of the BKV miRNA mimic and the resultant cDNA was serially diluted. A standard curve of mean Cq values, versus cDNA concentration, was used to extrapolate the concentration of BKV miRNA in patient samples (Figure 3). The limit of detection under these conditions corresponded to a Cq of <31 and 1000 molecules of bkv-miR-B1-5p.
Validation of bkv-miR-B1-5p detection in plasma from renal transplant recipients
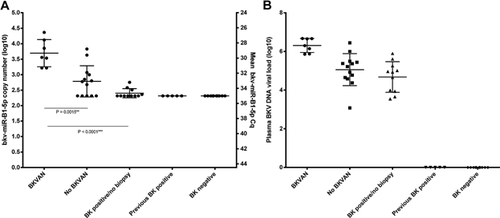
To determine if detection of bkv-miR-B1-5p was associated with BKVN we examined archived samples from renal transplant patients, which had previously been examined for BKV DNA by PCR because of deterioration of graft function. Stored blood samples were obtained from 46 patients in five groups: Group 1: patients with a positive BKV DNA PCR and biopsy-proven BKVN (BKVN) (n = 7); Group 2: patients with a positive BKV DNA PCR but no BKVN on biopsy (No BKVN) (n =13); Group 3: patients with detectable BKV DNA but in whom a renal biopsy was not undertaken (BK positive/no biopsy) (n = 11); Group 4: patients who were BKV DNA PCR positive but had become negative (previous BK positive, mean time to negative 12 months) at the time of this study (n = 5); and Group 5: a control group consisting of patients who were consistently negative for BKV DNA posttransplant until May 2013 (BK negative; n = 10). For patients who underwent renal biopsy, samples were selected that had been obtained closest in timing to the renal biopsy. Patients' characteristics in each group are summarized in Table 1. In keeping with the usual approach to management of patients with potential BKVN, there was a greater likelihood of undertaking renal biopsy in patients with higher levels of plasma BKV DNA and more impaired renal function.
As in the pilot study, we were able to detect bkv-miR-B1-5p in the plasma of patients who were positive for circulating BKV DNA and there was a significant correlation between bkv-miR-B1-5p and BKV DNA assays (Figure 4). By contrast, we were unable to detect bkv-miR-B1-5p in samples in which BKV DNA had not been detected. The highest levels of bkv-miR-B1-5p were apparent in patients with biopsy-proven BKVN. The Cq value and derived copy numbers (Figure 5A) of bkv-miR-B1-5p in the biopsy-proven BKVN (Group 1) were significantly higher than those with high plasma viral load but biopsy negative for BKVN (Group 2) and those with high plasma viral load who did not undergo biopsy (Group 3). Furthermore, there was no bkv-miR-B1-5p detected in serum from patients with very low circulating BKV DNA, control patients without BKV replication or those who had previously had BKV DNA detected in blood but were now negative. In keeping with previous studies, plasma BKV DNA viral load in patients with biopsy-proven BKVN was significantly higher than in patients without BKVN on renal biopsy (Figure 5B). Our data demonstrated a 10-fold higher mean expression of BKV miRNA in the blood of patients with biopsy-proven BKVN compared with patients positive for BKV DNA without evidence of BKVN on biopsy (p = 0.0015).
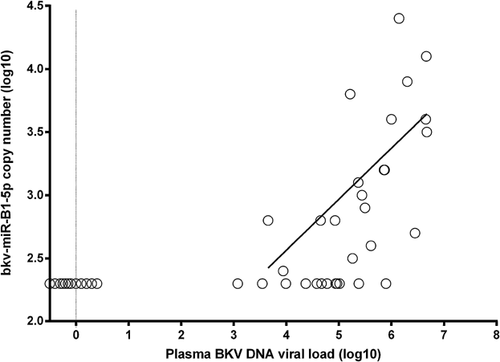
Given the unlikely possibility that other clinical features might influence the detection of BKV miRNA, we examined for correlations between the level of bkv-miR-B1-5p and other measurements (Table 2). However, while the Cq values of bkv-miR-B1-5p were strikingly inversely correlated with the BKV DNA levels (r = −0.766; p < 0.0001), variations in renal function, hemoglobin, platelets or the circulating levels of a human miRNA (miR-16) were not significantly associated with bkv-miR-B1-5p detection.
| BK PCR viral load | bkv-miR-B1-5p | hsa-miR-16 | ||||
|---|---|---|---|---|---|---|
| r | p | r | p | r | p | |
| eGFR (mL/min/1.73 m2) | −0.223 | 0.14 | 0.173 | 0.25 | −0.405 | 0.005 |
| Creatinine (µmol/L) | 0.187 | 0.21 | −0.114 | 0.45 | 0.377 | 0.010 |
| Hemoglobin (g/L) | −0.168 | 0.26 | 0.088 | 0.56 | −0.358 | 0.015 |
| Platelet count (×109/L) | −0.147 | 0.33 | −0.038 | 0.80 | −0.21 | 0.16 |
| BK PCR viral load (per mL) | – | – | −0.766 | <0.001 | 0.174 | 0.25 |
| bkv-miR-B1-5p | −0.766 | <0.001 | – | – | 0.109 | 0.47 |
| hsa-miR-16 | 0.174 | 0.25 | 0.109 | 0.47 | – | – |
- BKV, BK virus; Cq, quantification cycle; eGFR, estimated GFR; miRNA, microRNA; qPCR, quantitative polymerase chain reaction; sMDRD, simplified Modification of Diet in Renal Disease.
- Correlations between the Cq values of miRNAs hsa-miR-16 and bkv-miR-B1-5p with measurements of BKV DNA viral load, eGFR (sMDRD), creatinine, hemoglobin and platelet for n = 46 samples; qRT-PCR reactions with Cq values >35 or with poor amplification efficiency were treated as a negative result and assigned a Cq value of 35 in the analysis. Spearman's nonparametric correlation two-tailed p-value; significant p-values (<0.05) shown in bold.
Correlation between plasma bkv-miR-B1-5p, BKV DNA viral load and the presence of BKVN
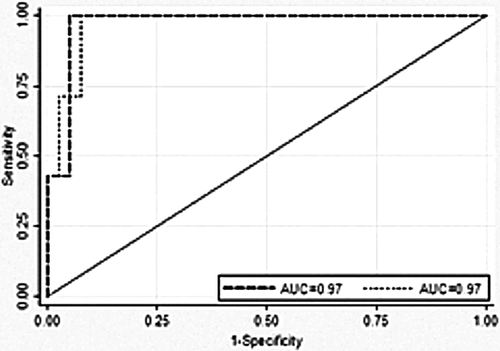
To examine the ability of bkv-miR-B1-5p and BKV DNA levels to determine the presence of BKVN, we undertook ROC analyses (Figure 6) on the 46 patients. The optimum cut-point for bkv-miR-B1-5p was at a predicted probability for biopsy-proven BKVN of 17.3%, which corresponded to a bkv-miR-B1-5p Cq of 31.9. This provided a sensitivity of 100.0% and a specificity of 94.9%. The area under the ROC was 0.97. The optimum cut-point for plasma DNA viral load was at a predicted probability for biopsy-proven BKVN of 7.7% corresponding to a plasma DNA viral load of 410 000. This provided a sensitivity of 100.0% and a specificity of 92.3%. The area under the ROC was 0.97. There was no significant difference between the C-statistic for bkv-miR-B1-5p and plasma BKV DNA viral load (p = 1.00). Utilizing the bkv-miR-B1-5p test with a Cq cutoff of 31.9 gave a positive-predictive value of 77.8% and negative-predictive value of 100% for BKVN, while utilizing plasma viral DNA load with a cut-point of 410 000 gave a positive-predictive value of 70.0% and negative-predictive value of 100% for BKVN. No patients with BK positivity who did not undergo renal biopsy had a Cq less than 31.9.
Discussion
We demonstrate for the first time that miRNAs encoded by BKV can be detected in blood and urine of patients with BKV infection. However, we were unable to detect these molecules in individuals without infection or in those no longer positive for circulating BKV DNA. The detection of high levels of bkv-miR-B1-5p in blood was strongly associated with high levels of BKV DNA and with biopsy-proven BKVN. These samples were examined retrospectively, and in keeping with the stability of these molecules, we were able to detect significant expression despite long-term freezer storage.
This study shows a very substantial difference between the levels of bkv-miR-B1-5p in patients with biopsy-proven BKVN and those with positive BKV DNA in the blood without biopsy evidence of nephropathy or those whose clinical features did not lead to biopsy. Furthermore, BKV miRNA is not detectable in patients without BKV replication. There was a strong correlation between the plasma levels of bkv-miR-B1-5p and BKV DNA determined by PCR. It remains to be established if BKV miRNA levels are solely indicative of high viral loads or have specificity for nephropathy.
In patients with severe chronic kidney disease, we recently found that circulating miRNAs were reduced in comparison to patients with normal renal function 23 suggesting that kidneys do not function in the clearance of circulating miRNAs. Given the potential for other clinical features to influence the detection of BKV miRNA, we examined for correlations between bkv-miR-B1-5p and other measurements. However, variations in renal function, hemoglobin or miR-16 levels were not significantly associated with bkv-miR-B1-5p detection. This is most in keeping with the levels of bkv-miR-B1-5p being associated with the severity of BKV replication.
However, this study has its limitations as it was a retrospective study of available archived blood specimens in a small number of patients. It is not capable of providing evidence that bkv-miR-B1-5p detection can perform as a potential diagnostic test for BKVN, nor that such miRNAs have a definite pathophysiological role. It does suggest the need for further research to determine whether in this and other viral infections the detection of virally encoded miRNAs can have diagnostic utility and distinguish latent from active replication. Evidence for this would require a large-scale study examining the detection of bkv-miR-B1-5p in blood and urine of renal transplant recipients. It could also address whether these molecules are detectable at differing stages of infection compared with existing DNA assays, whether they persist differently following remission or whether urine testing is useful. Furthermore, the half-life of BKV DNA and miRNA molecules may be very different.
The definitive diagnosis of BKVN can be made by renal allograft biopsy and histological findings on biopsy have been characterized and correlated with graft outcome 24. However, changes of BKVN can be focal or isolated to the medulla and missed on one-third of biopsies if only a single core is evaluated 24 and the histological finding of acute rejection can be similar to BKVN. The progression from BKV in the urine to BKV in the blood to nephropathy is often regarded as a stepwise transition and provides an opportunity for early detection and early intervention with reduction of immunosuppression 7. The recent American Society of Transplantation Infectious Diseases Guidelines recommended that in patients with sustained plasma BKV DNA loads of >104 copies/mL, a diagnosis of “presumptive BKVN” should be made in absence of demonstrable BKV replication in biopsies 25. However, a wide variety of primers, probes and standards for quantitative PCR assays are in use, which limits the usefulness of generalized quantitative viral load cutoffs in screening and diagnostic protocols. Furthermore, there is a broad range of sequence variation in different clinical isolates of BKV so that PCR assays vary in their ability to detect viral DNA 26 and can lead to false negative tests.
In this study, high levels of both BKV DNA and BKV miRNA are strongly predictive of BKVN. However, this study is not designed or powered to directly address whether BK miRNA detection has a better (or worse) predictive value in the noninvasive diagnosis of BKVN. Indeed, while the detection of BK miRNA had a good specificity for the presence of biopsy-proven BKVN, it appeared less sensitive for the detection of BKV DNA in the blood and assayed by this method would not usefully replace BK viral DNA screening protocols. Furthermore, the lower sensitivity of RT-PCR for RNA compared with DNA PCR may account for the correlation between higher plasma viral loads and BKV miRNA levels, and for the apparently high specificity of BKV miRNA levels in predicting BKVN.
This is the first demonstration that circulating BKV encoded miRNA bkv-miR-B1 is detectable in the plasma of patients with biopsy-proven BKVN and those with significant BKV DNA in the blood. It has also shown that bkv-miR-B1-5p levels are highest in patients with biopsy-proven BKVN. It suggests that circulating virally encoded miRNAs may act as indicators of severity of virus replication.
Acknowledgments
This study was supported by a seeding grant from the Flinders University Faculty of Health Sciences. We thank the patients and their clinicians for supporting this study.
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.