Argentinian clinical genomics in a leukodystrophies and genetic leukoencephalopathies cohort: Diagnostic yield in our first 9 years
Abstract
Introduction and objectives
Leukodystrophies and genetic leukoencephalopathies constitute a vast group of pathologies of the cerebral white matter. The large number of etiopathogenic genes and the frequent unspecificity on the clinical-radiological presentation generate remarkable difficulties in the diagnosis approach. Despite recent and significant developments, molecular diagnostic yield is still less than 50%. Our objective was to develop and explore the usefulness of a new diagnostic procedure using standardized molecular diagnostic tools, and next-generation sequencing techniques.
Materials and methods
A prospective, observational, analytical study was conducted in a cohort of 46 patients, evaluated between May 2008 and December 2016, with a suspected genetic leukoencephalopathy or leukodystrophy. A diagnostic procedure was set up using classical monogenic tools in patients with characteristic phenotypes, and next-generation techniques in nonspecific ones.
Results
Global diagnostic procedure yield was 57.9%, identifying the etiological pathogenesis in 22 of the 38 studied subjects. Analysis by subgroups, Sanger method, and next-generation sequencing showed a yield of 64%, and 46.1% respectively. The most common pathologies were adrenoleukodystrophy, cerebral autosomal-dominant arteriopathy with subcortical infarcts (CADASIL), and vanishing white matter disease.
Conclusions
Our results confirm the usefulness of the proposed diagnostic procedure expressed in a high diagnostic yield and suggest a more optimal cost-effectiveness in an etiological analysis phase.
1 INTRODUCTION
Genetic leukodystrophies and leukoencephalopathies constitute a vast group of neurological diseases that affect all age groups, with the involvement of cerebral white matter as a common denominator. Under the term leukodystrophies are those genetic disorders induced by primary damage of glial cells or myelin of the central nervous system, with or without involvement of the peripheral nervous system. On the other hand, genetic leukoencephalopathies include hereditary disorders of the white matter that cannot be included in the term leukodystrophies, such as those entities with primary involvement of the gray matter or systemic pathologies with secondary neurological involvement (example: mitochondrial diseases with hepatic, muscular, or cardiac involvement and additional commitment of central nervous white matter) (Vanderver et al., 2015). Between 1990 and 2010, many of the genes linked to these pathologies were identified. In the last few years, thanks to the incorporation of massive sequencing techniques, a noticeable expansion in the knowledge of the genetic causes of white matter diseases was made possible, thus allowing the identification of more than 100 leukodystrophies and genetic leukoencephalopathies.
Despite these significant advances, the molecular diagnostic yield is still less than 50% (Kevelam et al., 2016). The nonspecificity of the symptoms, the clinical superposition between one entity and another, the heterogeneous presentation, the limited existence of biochemical markers, and the little guidance in the findings in neuroimaging several of the disorders added to the high number of genes involved generate marked difficulties when making a diagnostic approach, even for medical specialists in the field. The need arises to find an ideally simple, economic, and reproducible procedure that allows us to reach the etiological diagnosis in a greater number of cases, which would have profound implications in the clinical follow-up, family counseling, and eventual therapeutic behavior, depending on the case. On the other hand, although there have been isolated reports of individual patients with leukodystrophies and genetic leukoencephalopathies, and small series of cases in the country and region (Amorosi et al., 2012; Arroyo, Grippo, Taratuto, Duffau, & Chamoles, 1991; de Kremer et al., 2002; Guarnaschelli & Sotelo, 2017; Rodríguez-Quiroga et al., 2015), there has not been a systematic analysis evaluating the diagnostic performance of a comprehensive molecular diagnosis approach in the field of these disorders in Argentina.
Therefore, the objectives of our work were (1) to characterize and stratify a population of adult and pediatric patients with leukodystrophies and genetic leukoencephalopathies, and (2) to develop and evaluate the diagnostic performance of a clinical-molecular procedure, which used classical and next-generation sequencing (NGS) techniques in a prospective cohort of patients with neurogenetic disorders.
2 MATERIALS AND METHODS
2.1 Patients: Diagnostic procedure
A prospective, observational, and descriptive cohort study was conducted, including adult and pediatric patients enrolled at the Neurogenetic Clinic of the J.M. Ramos Mejía Hospital in Buenos Aires, from May 2008 to December 2016, with a clinical diagnosis of leukodystrophy or genetic leukoencephalopathy, based on the definition of both concepts by Vanderver et al. (2015). All the patients, prior to their participation in the study, provided informed consent through a form approved by the Ethics Committee of the institution, and in the case of minors, the consent was given by their parents. Demographic, family history, clinical, and paraclinical data were obtained.
For the selection of the molecular tests to be performed in each patient, the clinical characteristics, biochemical markers, and neuroimaging findings of each individual case were considered. In those who presented with a phenotype characteristic of a pathology linked to a single candidate gene and capable of being analyzed by the Sanger method, such as, but not limited to, X-linked adrenoleukodystrophy, metachromatic leukodystrophy, cerebral autosomal-dominant arteriopathy with subcortical infarcts (CADASIL), and hereditary diffuse leukoencephalopathy with spheroids, we proceeded to the sequencing of the etiopathogenic gene using the technique described for ABCD1, ARSA, NOTCH3, and CSF1R, respectively. In cases with nonspecific phenotype and/or complex, genetic heterogeneity, or candidate gene not amenable to analysis by the Sanger method, a multigenic panel was used with the inclusion of more than 100 genes linked to leukodystrophies and genetic leukoencephalopathies, with NGS techniques. Unfortunately, funding did not permit multigene panel testing in those subjects (n = 9) for whom the Sanger method did not confirm an etiology (Figure 1; Supplementary Table 1).
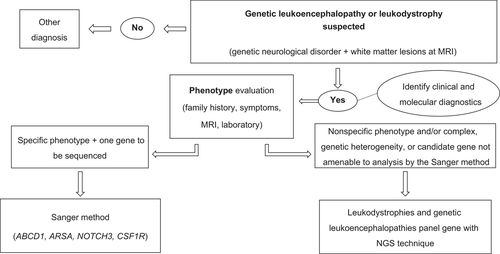
2.2 Molecular studies
2.2.1 Genomic DNA purification
Two millimeters of whole blood were obtained from each patient by venipuncture. Total genomic DNA was purified using commercial systems, following the manufacturer's instructions. This was kept anonymized until further processing.
2.2.2 Sequencing of individual genes by the Sanger technique
Primers were designed for the coding regions of the ABCD1 gene (*300371) using NCBI NM_000033.3 on GRCh37 (hg19) as the reference sequence. The 10 exons, including the flanking regions of each exon, were amplified by “polymerase chain reaction” (PCR) and sequenced by the Sanger method, as described in Boehm, Cutting, Lachtermacher, Moser, & Chong (1999).
For the analysis of the ARSA gene (*607574), the reference of GenBank NM_000487.5 was followed, based on GRCh37 (hg19). The adjacent coding and intronic regions of the eight exons were amplified by PCR using oligonucleotides or primers designed for that purpose and subsequent sequencing by the Sanger method (conditions available as required).
For the analysis of the NOTCH3 gene (*600276), the NCBI NM_000435.2 was used on the reference sequence GRCh37 (hg19) and the exons 3-4-5-6-11-12-18-19-20 were studied, while for the CSF1R gene analysis (*221820), based on NCBI NM_005211.3, the coding region and exon–intron junctions of exons 18, 19, and 20 were examined; in both cases, the same methodology to that already specified above was used (conditions available as required).
For the analysis of the GFAP gene (*137780) and EIF2B5 gene (*603945), the NCBI NM_000435.2 and NCBI NM_003907.2 were used on the reference sequence GRCh37 (hg19), respectively, sequencing the coding region and exon–intron junctions following the Sanger method (conditions available as required).
2.2.3 Multigene panel sequencing
The coding fraction of 4813 genes was enriched in solution following standardized procedures recommended by the manufacturer using the TruSight One Sequencing Panel commercial kit (Illumina, San Diego, CA). Later, in batches of three samples, sequencing runs were performed on Illumina MiSeq equipment, achieving average coverage greater than 70X, following standardized procedures recommended by the manufacturer. A total of three sequencing runs were performed for a total analysis of nine samples. Four other patients underwent similar procedures but in commercial genomic diagnostic laboratories (methods reported in individual study reports). As mentioned below, the clinical interpretation was based on the scrutiny of a subset of genes associated with white matter disorders (virtual panel analysis).
2.3 Bioinformatic analysis
The product of the sequencing runs was aligned with the reference sequence of the human genome of the National Center for Biotechnology Information of the National Institutes of Health of the United States version 37/hg19 using the BWA tool (version 0.5.9, requirement) (Li & Durbin, 2009). Subsequently, single-nucleotide variants and small insertions/deletions were identified, using the tools Picard (version 1.59) and GATK (version 3.6, parameters used available upon request) (Van der Auwera et al., 2013). From a search of genes reported as causing leukodystrophies and genetic leukoencephalopathies in OMIM and HGMD, a virtual multigenic panel (Annex I) of the pathologies under study was constructed, which was useful for the analysis and clinical interpretation of the variant files obtained from sequencing runs, using procedures developed by our group and described in Córdoba et al. (2018).
3 RESULTS
3.1 Cohort
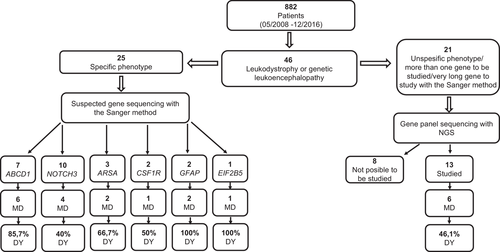
Of the 882 patients evaluated in the Neurogenetic Clinic of our hospital from May 2008 to December 2016, we included 46 subjects with diagnostic presumptions of leukodystrophy or genetic leukoencephalopathy.
-
a group of patients with a phenotype characteristic of a pathology linked to a single candidate gene, and capable of being analyzed by the Sanger method, in which individual sequencing was carried out using the technique described; such is the case of X-linked adrenoleukodystrophy, metachromatic leukodystrophy, CADASIL, vanishing white matter disease, Alexander's disease, and diffuse hereditary leukoencephalopathy with axonal spheroids and pigmented cells, and
-
a group of patients with nonspecific phenotype and/or complex, genetic heterogeneity, or candidate genes not amenable to analysis by the Sanger method, who were evaluated through extended gene sequencing with next-generation techniques.
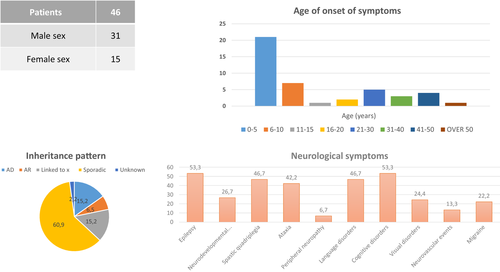
The methodology used and the cohort studied by strata are represented in Figure 2 with the diagnostic yield obtained in each case. The demographic and clinical characteristics of the included patients are shown in Figure 3. The details of the clinical presentation, radiology, family history, diagnostic presumption, and method of molecular study used in each patient, are presented in Table 1.
| Patient number | Gender | Age of onset (years) | Clinical | Brain MRI | Family history inherited pattern | Clinical diagnosis | Gene sequencing method |
|---|---|---|---|---|---|---|---|
| 1 | M | 10 | Epilepsy: spastic quadriparesis, language disorder, evolutive, Addison syndrome, increased VLCFA | White matter images: hyperintense in T2 and hypointense in T1, bilateral parietal and occipital, Gd (+) |
Six cousins affected X- linked |
X-linked adrenoleukodystrophy | Sanger method: ABCD1 gene |
| 2 | M | 9 | Epilepsy, spastic quadriparesis, language and cognitive disorder, evolutive, Addison syndrome increased VLCFA | White matter images, hyperintense in T2 and hypointense in T1, bilateral parietal and occipital |
Grandfather and brother affected X-linked |
X-linked adrenoleukodystrophy | Sanger method: ABCD1 gene |
| 3 | M | 7 | Epilepsy, spastic quadriparesis, language and cognitive disorder, evolutive. Addison syndrome, increased VLCFA | White matter images, hyperintense in T2 and hypointense in T1, bilateral parietal and occipital, Gd (+) |
Sister carrier, X-linked |
X-linked adrenoleukodystrophy | Sanger method: ABCD1 gene |
| 4 | M | 6 | Epilepsy, spastic quadriparesis, language and cognitive disorder, evolutive, Addison syndrome, increased VLCFA | White matter images, hyperintense in T2 and hypointense in T1, bilateral frontal, parietal and occipital, Gd (+) | Uncle affected, sister carrier, X-linked | X-linked adrenoleukodystrophy | Sanger method: ABCD1 gene |
| 5 | M | 18 | Learning disorders, pyramidal syndrome, periferic neuropathy, evolutive, increased VLCFA | White matter images, hyperintense in T2 and hypointense in T1, bilateral parietal and occipital with involvement of brainstem and cerebellum | Brother with increased VLCFA | X-linked adrenoleukodystrophy | Sanger method: ABCD1 gene |
| 6 | M | 20 | Peripheral neuropathy, pyramidal syndrome, language and cognitive disorder, evolutive, Addison syndrome, increased VLCFA | Right periventricular white matter image hyperintense in T2, tractography: decrease in the fibers of the pyramidal path |
Sister carrier. X-linked |
X-linked adrenoleukodystrophy | Sanger method: ABCD1 gene |
| 7 | M | 3 | Epilepsy, without symptoms for 13 years, then ataxia, spastic quadriparesis, language disorder, evolutive, increased VLCFA | White matter images, hyperintense in T2 and hypointense in T1, bilateral parietal and occipital |
Mother with peripheral neuropathy, X-linked |
X-linked adrenoleukodystrophy | Sanger method: ABCD1 gene |
| 8 | F | 47 | TIA, migraine with aura, mood disorders |
Bihemispheric white matter images hyperintense in T2, with temporal pole involvement |
Autosomal dominant |
CADASIL | Sanger method: NOTCH3 gene |
| 9 | M | 34 | TIA, migraine without aura | Bihemispheric white matter images hyperintense in T2, with temporal pole involvement | Sporadic | CADASIL | Sanger method: NOTCH3 gene |
| 10 | F | 32 | Epilepsy, migraine with aura | Bihemispheric white matter images hyperintense in T2, with temporal pole involvement |
Autosomal dominant |
CADASIL | Sanger method: NOTCH3 gene |
| 11 | M | 27 | TIA | Bihemispheric white matter images hyperintense in T2, with temporal pole involvement |
Autosomal dominant |
CADASIL | Sanger method: NOTCH3 gene |
| 12 | F | 30 | Stroke, migraine without aura, epilepsy, cognitive disorder | Bihemispheric white matter images hyperintense in T2, with temporal and frontal pole involvement |
Autosomal dominant |
CADASIL | Sanger method: NOTCH3 gene |
| 13 | F | 21 | Stroke | Bihemispheric white matter images hyperintense in T2, with supra and infratentorial involvement. | Sporadic | CADASIL | Sanger method: NOTCH3 gene |
| 14 | F | 40 | Migraine, cognitive disorder | Bihemispheric white matter images hyperintense in T2, with temporal pole involvement | Sporadic | CADASIL | Sanger method: NOTCH3 gene |
| 15 | M | 44 | Migraine, arterial hypertension | Bihemispheric white matter images hyperintense in T2, with temporal pole and external capsule involvement |
Autosomal dominant |
CADASIL | Sanger method: NOTCH3 gene |
| 16 | M | 46 | Migraine, cognitive disorder | Bihemispheric white matter images hyperintense in T2, with temporal pole and external capsule involvement |
Autosomal dominant |
CADASIL | Sanger method: NOTCH3 gene |
| 17 | M | 43 | Stroke, migraine with aura, arterial hypertension | Bihemispheric white matter images hyperintense in T2, with temporal pole and external capsule involvement | Sporadic | CADASIL | Sanger method: NOTCH3 gene |
| 18 | M | 3 | Epilepsy, intellectual disability, low weight and short stature, macrocephaly | Bilateral periventricular white matter images, hyperintense in T2, with infratentorial and spinal cord involvement | Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 19 | M | 1 | Epilepsy, intellectual disability, macrocephaly | Bihemispheric white matter images, hyperintense in T2, with frontal lobe involvement | Sporadic | Alexander disease | Sanger method: GFAP gene |
| 20 | F | 0 |
Epilepsy, intellectual disability, macrocephaly, spastic quadriparesis, ataxia |
Bihemispheric white matter images, hyperintense in T2, with frontal lobe involvement | Sporadic | Alexander disease | Sanger method: GFAP gene |
| 21 | F | 1 |
Epilepsy, intellectual disability, microcephaly, spastic quadriparesis, ataxia, strabismus and nystagmus, hypoacusis, low weight and short stature, dysmorphism, hypogonadism, hypoparathyroidism |
Diffuse cortical atrophy, hyperintensity of lenticular nucleus in T1, bilateral periventricular images hyperintense in T2 hypointense in T1, corpus callosum atrophy |
Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 22 | M | 6 | Ataxia, cognitive and conductal disorder, visual involvement and nystagmus, hypoacusis |
Bilateral periventricular images hyperintense in T2 hypointense in T1, corpus callosum and brainstem atrophy |
Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 23 | M | 1 | Epilepsy, intellectual disability, ataxia, spastic quadriparesis, hypodontia, dysmorphia | Bihemispheric white matter images hyperintense in T2, hypointense in T1 | Sporadic | 4 “H” leukodystrophy | Plan: panel gene sequencing with NGS technique (genetic heterogeneity) (this patient could not be studied) |
| 24 | M | 10 | Epilepsy, intellectual disability, ataxia, spastic quadriparesis evolutive, decreased in arylsulfatase A activity and increased in urine sulfatides |
Diffuse cortical atrophy, bilateral periventricular images hyperintense in T2 hypointense in T1, corpus callosum atrophy |
Autosomal recessive | Metachromatic leukodystrophy | Sanger method: ARSA gene |
| 25 | M | 1 | Intellectual disability, spastic quadriparesis evolutive, decreased in arylsulfatase A activity and increased in urine sulfatides | Bilateral periventricular images hyperintense in T2 hypointense in T1 | Sporadic | Metachromatic leukodystrophy | Sanger method: ARSA gene |
| 26 | M | 1 | Epilepsy, intellectual disability, ataxia, spastic quadriparesis evolutive, decreased in arylsulfatase A activity and increased in urine sulfatides | Bilateral periventricular images hyperintense in T2 hypointense in T1 with infratentorial and corpus callosum involvement | Sporadic | Metachromatic leukodystrophy | Sanger method: ARSA gene |
| 27 | M | 27 | Evolutive, spastic quadriparesis, saccadic eyes movement disorder, cognitive impairment | Asymmetric bihemispheric white matter images, hyperintense in T2, with frontal lobe involvement |
Autosomal dominant |
Hereditary diffuse leukoencephalopathy with spheroids and pigmented cells | Sanger method: CSF1R gene |
| 28 | F | 71 | Ataxia, cognitive disorder, evolutive | Asymmetric bihemispheric white matter images, hyperintense in T2, with frontal lobe involvement | Sporadic | Hereditary diffuse leukoencephalopathy with spheroids and pigmented cells | Sanger method: CSF1R gene |
| 29 | F | 1 |
Intellectual disability, epilepsy, macrocephaly, Ataxia |
Bihemispheric hypomyelination with subcortical temporal cysts | Unknown | Megalencephalic leukodystrophy with subcortical cysts | Panel gene sequencing with NGS technique (this gene is not candidate to individual sequencing) |
| 30 | F | 15 | Epilepsy, left hemiparesis, pyramidal syndrome, ataxia, cognitive disorder, evolutive | Asymmetric bihemispheric white matter images, hyperintense in T2 | Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 31 | F | 3 | Ataxia, status epilepticus and posterior progressive encephalopathy, spastic quadriparesis, microcephaly, Primary amenorrhea and absence of secondary sexual characteristics | Bihemispheric white matter images, hyperintense in T2 with tendency to cysts formations, diffuse atrophy | Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 32 | F | 5 | Brain injury then ataxia and posterior recovery, acute left hemiparesis, migraine, epilepsy, premature ovarian failure | Asymmetric bihemispheric white matter images, hyperintense in T2 with tendency to cysts formations, diffuse atrophy | Sporadic | Vanishing white matter disease | Sanger method: EIF2B5 gene, exon 7 |
| 33 | M | 2 | Brain injury then ataxia and posterior recovery, other episode of brain injury then left hemiparesis and posterior partial recovery, pyramidal syndrome | Bihemispheric white matter images, hyperintense in T2 with tendency to cysts formations, diffuse atrophy | Sporadic | Vanishing white matter disease | Panel gene sequencing with NGS technique (genetic heterogeneity) |
| 34 | M | 4 | Evolutive cognitive disorder | Bihemispheric white matter images, hyperintense in T2 with tendency to cysts formations | Sporadic | Vanishing white matter disease | Plan: panel gene sequencing with NGS technique (genetic heterogeneity) (this patient could not be studied) |
| 35 | M | 1 | Intellectual disability, spastic quadriparesis, visual disorders | Bihemispheric white matter images, hyperintense in T2 with tendency to cyst formation, diffuse atrophy | Sporadic | Vanishing white matter disease | Plan: panel gene sequencing with NGS technique (genetic heterogeneity) (this patient could not be studied) |
| 36 | M | 3 | Epilepsy, spastic quadriparesis, ataxia, cognitive disorder, hypoacusis, evolutive | Bihemispheric white matter images, hyperintense in T2 with corpus callosum and infratentorial involvement | Sporadic | Unspecific phenotype | Plan: panel gene sequencing with NGS technique (this patient could not be studied) |
| 37 | M | 33 | Visual disorders, transient focal neurological disorders, migraine, hypoacusis | Bihemispheric white matter images, hyperintense in T2 with tendency to cysts formations | Sporadic | Unspecific phenotype | Plan: panel gene sequencing with NGS technique (this patient could not be studied) |
| 38 | M | 0 | Epilepsy, intellectual disability, spastic quadriparesis, ataxia, cognitive disorder | Bihemispheric white matter images, hyperintense in T2 with supra and infratentorial involvement | Sporadic | Unspecific phenotype | Plan: panel gene sequencing with NGS technique (this patient could not be studied) |
| 39 | F | 4 | Epilepsy, spastic quadriparesis, language disorder, evolutive, systemic lupus erythematosus | Bihemispheric white matter images, hyperintense in T2 with basal ganglia calcification | Sporadic | Aicardi-Goutières | Plan: panel gene sequencing with NGS technique (this patient could not be studied) |
| 40 | M | 40 | Episodic vertigo, hypothyroidism | Bihemispheric white matter images, hyperintense in T2 with supra and infratentorial involvement | Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 41 | M | 1 | Epilepsy, intellectual disability, spastic quadriparesis, cognitive disorder, microcephaly | Bilateral periventricular white matter images, hyperintense in T2 | Autosomal recessive | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 42 | F | 28 | Epilepsy, ataxia, cognitive disorder, evolutive, premature ovarian failure, hypothyroidism | Bihemispheric white matter images, hyperintense in T2 with supra and infratentorial involvement | Sporadic | Unspecific phenotype | Plan: panel gene sequencing with NGS technique (this patient could not be studied) |
| 43 | M | 1 | Epilepsy, intellectual disability, spastic quadriparesis, cognitive and visual disorders | Bihemispheric white matter images, hyperintense in T2 with tendency to cysts formations | Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 44 | M | 10 | Migraine, cognitive disorder, pyramidal syndrome | Bihemispheric white matter images, hyperintense in T2 | Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 45 | M | 2 | Progressive spastic paraparesis, hollow foot | Supratentorial bihemispheric white matter images, hyperintense in T2 | Sporadic | Unspecific phenotype | Panel gene sequencing with NGS technique |
| 46 | F | 1 | Intellectual disability, evolutive, ataxia, spastic quadriparesis | Bihemispheric white matter images, hyperintense in T2 with corpus callosum involvement | Autosomal recessive | Unspecific phenotype | Panel gene sequencing with NGS technique |
- Abbreviations: MRI, Magnetic resonance imaging; M, male; F, female; VLCFA, very long-chain fatty acids; TIA, transient ischemic attack; CADASIL, cerebral autosomal-dominant arteriopathy with subcortical infarcts; Gd (+), gadolinium positive.
3.2 Diagnostic yield of the comprehensive approach
From this study methodology and using the proposed diagnostic procedure, we arrived at the molecular diagnosis that confirmed the genetic etiology in 22 subjects of the 38 studied (57.9%). It was not possible to perform molecular studies in eight patients that were not available for blood testing. The details of the recognized mutations are summarized in Supplementary Table 2.
In the case of patients with suspected X-linked adrenoleukodystrophy, based on family history, very long-chain fatty acids, endocrine comorbidity with adrenal insufficiency, and suggestive radiological abnormalities, the ABCD1 gene was sequenced by the Sanger method, confirming the molecular diagnosis in six of the seven patients evaluated (85.7%).
In three patients, there was a diagnostic suspicion of metachromatic leukodystrophy based on the biochemical profile, which shows a decrease in the plasmatic activity of the enzyme arylsulfatase A and an increase in the excretion of urinary sulfatides. The sequencing of the ARSA gene showed the presence of pathogenic variants in homozygosis in two of them (66.7%). In the patient in whom only one heterozygous variant was found, the occurrence of a structural variant in the other allele or the presence of a pathogenic variant in areas that are not sequenced by the Sanger method are possible.
The presumptive diagnosis of CADASIL was based on the occurrence of acute neurovascular events, migraine, or cognitive impairment in individuals under 50 years old, with or without family history compatible with autosomal-dominant inheritance and associated with neuroimaging with multifocal involvement of cerebral white matter. The diagnosis was confirmed by the finding of pathogenic variants in the NOTCH3 gene in four of the 10 patients studied (40%).
In the cases of a progressive adult-onset neurological disorder, characterized by cognitive-behavioral impairment and asymmetric pyramidal motor deficit associated with magnetic resonance imaging (MRI) showing involvement of cerebral white matter with frontoparietal predominance, with or without an autosomal-dominant inheritance pattern, we suspected diffuse hereditary leukoencephalopathy with axonal spheroids and pigmented giant cells. The sequencing of the CSF1R gene revealed a pathogenic variant in heterozygosis in one of the two patients evaluated with these characteristics (50%).
In 13 patients, we proceeded to study a multigenic panel by massive sequencing in parallel, with next-generation techniques through the analysis of more than 100 genes linked with leukodystrophies and genetic leukoencephalopathies. All of them presented variable clinical forms, without radiological signs or neurometabolic studies suggestive of any particular entity, or because of genetic heterogeneity or the size of the candidate gene made the individual approach difficult. Molecular diagnosis was obtained in six of them (46.1%).
3.3 Illustrative cases
3.3.1 Case No. 1 (Patient No. 1)
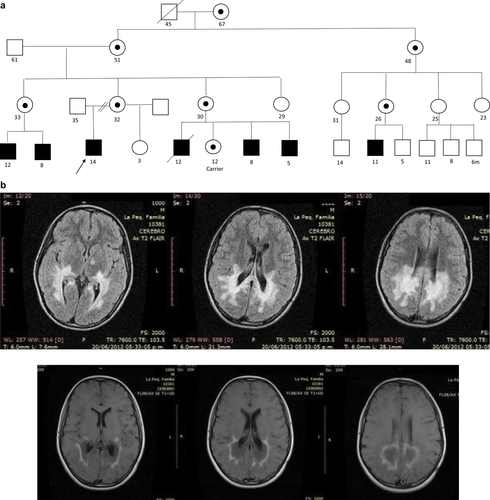
A male patient, at 10 years old, presented with convulsions, progressive neurologic deterioration manifesting as pyramidal motor compromise, changes in skin color, visual symptoms, and a family history suggestive of X-linked inheritance (Figure 4a). Neurological examination revealed severe phonological deglutition disorder, emission of incomprehensible sounds, bilateral visual acuity affectation with homonymous hemianopsia and convergent strabismus, spastic quadriparesis with pyramidal signs, and cervical dystonic posture. MRI of the brain showed confluent images in posterior periventricular white matter, hyperintense in T2 and spin echo sequences enhanced in T2, hypointense in T1, with peripheral enhancement after contrast administration (Figure 4b). Laboratory findings included increased adrenocorticotrophin and decreased cortisol compatible with adrenal insufficiency, and a neurometabolic study indicated an increase in very long-chain fatty acids. With presumptive diagnosis of X-linked adrenoleukodystrophy, the ABCD1 gene was sequenced, and c.320T > C variant (p.Leu107Pro) was detected in exon 1, classifiable as pathogenic and previously reported in the literature (Figure 4c; Korenke et al., 1997; Krasemann, Meier, Korenke, Hunneman, & Hanefeld, 1996).
3.3.2 Case No. 2 (Patient No. 30)
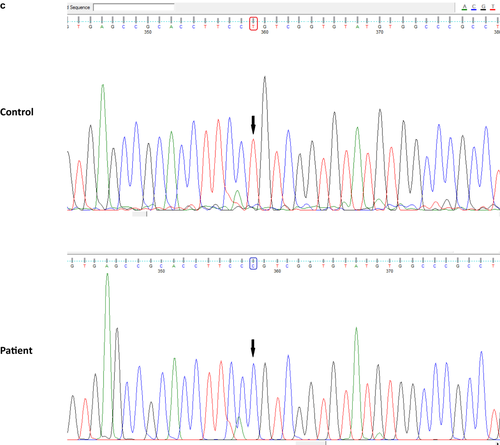
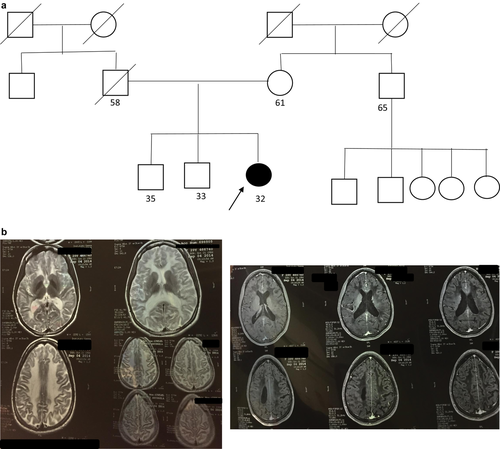
A 32-year-old female patient had presented at 15 years of age with progressive neurological symptoms, characterized by focal seizures, ataxia, motor deficit in the left side of the body, and cognitive impairment, with no family history (Figure 5a). A metabolic laboratory without relevant findings, and brain MRI with confluent lesions in bilateral periventricular and subcortical white matter, hyperintense in T2 and hypointense in T1, with tractography showed an asymmetric bilateral pyramidal pathway compromise (Figure 5b). Given that the phenotype did not approach a defined clinical-radiological syndrome, a multigenic panel sequencing was resorted to using next-generation techniques. A pathogenic variant in homozygosis was identified in exon 3 of the EIF2B3 gene (*606273): NM_001261418: c.260C > T (p.A87V), which has been previously reported in the literature as pathogenic (van der Knaap et al., 2002), concluding in the diagnosis of vanishing white matter disease.
4 DISCUSSION
In the present work, a pediatric and adult population of patients with genetic leukoencephalopathies and leukodystrophies was characterized, obtaining a genetic diagnosis in a significant number of individuals. This has been possible because of the development of a diagnostic procedure by means of which the molecular study path is addressed according to the presumption that arises from the phenotypic evaluation.
Although these entities are characterized by a marked clinical heterogeneity and involve a high number of candidate genes (Lynch et al., 2017), the identification of the causal defect was achieved in 22 of the 38 patients studied. Our results confirm the usefulness of the proposed diagnostic procedure, expressed by the significant diagnostic yield (57.9%). Noteworthy, the yield for Sanger sequencing in the CADASIL phenotype was 40%, whereas the yield for the remaining phenotypes was higher at 80%.
In patients with an uncharacteristic and/or complex phenotype, the use of a directed capture of genes through the NGS technology reaffirmed the performance of these techniques for the diagnostic approach in these pathologies (Wang et al., 2016). This constituted a quick and effective instrument, allowing us to arrive at a definite etiology even when the clinical manifestation is not eloquent (Gordon, Letsou, & Bonkowsky, 2014), reducing costs by shortening the long process of complementary methods of study that patients are often subjected to (Córdoba, González Morón, Rodríguez-Quiroga, & Kauffman, 2014).
This work covers the largest series of patients affected by genetic diseases of the cerebral white matter reported in Argentina, with the inclusion of clinical and molecular data. In relation to similar publications in other countries, we found similarities in clinical manifestations and in the genes studied, with individual differences in the frequency of each of the pathologies, probably because of the different epidemiological characteristics of the populations evaluated (Lynch et al., 2017; Stellitano, Winstone, van der Knaap, & Verity, 2016). Likewise, there was a greater proportion of adult patients because of the high number of people belonging to this age group being evaluated at our center, which is reflected in the diagnostic approach to pathologies characteristic of this group, such as CADASIL, and diffuse hereditary leukoencephalopathy with axonal spheroids and pigmented cells.
In other cohorts that studied populations of similar ages, the suspicion based on the clinical-radiological characterization was similar, with higher frequency of adult-onset vanishing white matter disease, and other pathologies such as leukodystrophy with ovarian failure related to the mutation in the AARS2 gene, which we did not have in our series (Lynch et al., 2017). The sequence of the NOTCH3 gene confirmed the diagnosis in only four of the patients studied with suspected CADASIL, unlike other reports where the diagnostic yield was higher (Chen et al., 2017). In our particular case, we recognize the laxity in the selection criteria, as we studied those patients who met at least one clinical and one radiological criterion described by Davous (1998) without being strict in the sum of 15 or more points on the diagnostic probability scale of CADASIL proposed by Pescini, Nannucci, & Pantoni (2013), there is a better performance in the molecular study of the NOTCH3 gene.
On the other hand, with respect to the patients studied with suspicion of X-linked adrenoleukodystrophy, in a single patient no pathogenic variants were detected in the ABCD1 gene, presuming that they may be found in noncoding regions of the gene or because of the presence of a deletion that escapes detection by the Sanger method. A similar case occurs in the patient with suspected metachromatic leukodystrophy in whom a confirmatory molecular diagnosis could not be reached because a single heterozygous pathogenic variant was detected. Therefore, it is believed that there may be another variant in noncoding regions or structural anomalies not identifiable by this method, requiring additional techniques such as MLPA assays not performed by our group. Finally, the case with suspicion of diffuse hereditary leukoencephalopathy with axonal spheroids and pigmented cells that we could not confirm, corresponded to a patient with a nonconclusive clinical phenotype, with difficulty in the subsequent follow-up.
The illustrated cases are paradigmatic. The first patient with phenotypic characteristics allowed suspicion of a particular entity, leading to the sequencing of an individual gene by classical techniques, with a high probability of achieving a definitive etiological diagnosis through a single molecular study. The second case represents a sufficiently complex phenotype to require a broader approach, feasibly to be analyzed through a next-generation technique as a directed panel of candidate genes, which in a single step allowed obtaining a molecular diagnosis, confirming a defined pathological entity, reducing time and costs (Richards, Korgenski, Taft, Vanderver, & Bonkowsky, 2015). The configuration of a diagnostic procedure allows the treating physician to have a better approach to this complex group of pathologies, optimizing the diagnostic resources.
The access to an etiological diagnosis derives in medical actions that originate from it, through the early detection or monitoring of the different comorbidities (Parikh et al., 2015). The possibility of providing adequate genetic counseling through the guidance on risk of recurrence is equally important, and, most of all, the obtaining of a greater proportion of molecular diagnoses expands the knowledge of the clinical spectrum of each genetic leukoencephalopathy, configuring a registry of the individualized causal defect for future therapeutic opportunities in current development. Moreover, comprehensive diagnostic approaches, such as the one used by our group, reduces the time that these patients must wait before getting a diagnosis thereby ending odysseys of many years, impacting in disorders such as ALD-X on their medical management, and optimizing the genetic counseling for these families. However, we must keep working on new methods that allow a rapid diagnosis of the genetic conditions of the cerebral white matter to reduce the individual and family impact.
The vast amount of knowledge that has emerged in recent years about the genes linked to leukodystrophies and genetic leukoencephalopathies will provide genomics an increasingly essential role as a tool for diagnostic use, which will lead to the growing need to develop therapeutic strategies to avoid the significant morbidity and mortality of these pathologies.
Despite the notorious advances achieved with the increasing use of massive sequencing techniques, a high percentage of affected individuals remain without genetic diagnosis. It is necessary to have larger groups of patients through collaborative work between the different groups of studies, to have a deeper and more extensive knowledge of these conditions. Research in the molecular diagnosis of genetic leukoencephalopathies and leukodystrophies continues to be a great challenge in this new era where knowledge and promising therapeutic advances evolve daily.
ACKNOWLEDGMENTS
We thank the patients and families for their support and collaboration, and Mrs. Verónica de Pablo, who coordinates the “Lautaro te Necesita” Foundation, for her valuable and inspiring daily work with children and families, as well as her trust in each one of the members of our team. This study was supported by the “Lautaro te Necesita” Foundation, providing the technical resources used through the gene panel implemented in the present work. This study was funded by a grant from the Ministry of Science and Technology of Argentina.
AUTHOR CONTRIBUTIONS
M.K. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. The study concept and design were performed by L.C., P.V., and M.K. Acquisition of data was done by L.C., A.M., H.A., N.S., M.C., and N.M. Analysis and interpretation of data was done by L.C., N.M., S.R.-Q., D.G.-M., J.R., and P.V. Manuscript preparation was performed by all authors.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
ETHICAL APPROVAL
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
INFORMED CONSENT
Informed consent was obtained from all individual participants included in the study.




