A genome-wide association study reveals a quantitative trait locus for days open on chromosome 2 in Japanese Black cattle
Summary
Days open (DO), which is the interval from calving to conception, is an important trait related to reproductive performance in cattle. To identify quantitative trait loci for DO in Japanese Black cattle, we conducted a genome-wide association study with 33 303 single nucleotide polymorphisms (SNPs) using 459 animals with extreme DO values selected from a larger group of 15 488 animals. We identified a SNP on bovine chromosome 2 (BTA2) that was associated with DO. After imputation using phased haplotype data inferred from 586 812 SNPs of 1041 Japanese Black cattle, six SNPs associated with DO were located in an 8.5-kb region of high linkage disequilibrium on BTA2. These SNPs were located on the telomeric side at a distance of 177 kb from the parathyroid hormone 2 receptor (PTH2R) gene. The association was replicated in a sample of 1778 animals. In the replicated population, the frequency of the reduced-DO allele (Q) was 0.63, and it accounted for 1.72% of the total genetic variance. The effect of a Q-to-q allele substitution on DO was a decrease of 3.74 days. The results suggest that the Q allele could serve as a marker in Japanese Black cattle to select animals with superior DO performance.
Days open (DO), the time from parturition until successful insemination, is a valuable index reflecting the efficiency of resuming the oestrus cycle after parturition and conception. Ideally, a cow produces a single calf each year (365 days). For a 365-day calving interval, the DO should not exceed 80–85 days. However, the recent average DO is 115 days for Japanese Black cattle (Wagyu Registry Association 2011). Thus, farmers aim to reduce the DO to improve reproductive performance and profitability; however, the genetic factors affecting DO in Japanese Black cattle remain unknown.
Quantitative trait loci (QTL) analyses have identified loci associated with DO and DO-related traits in cattle (Schulman et al. 2008; Hawken et al. 2012). However, these QTL have not been replicated, nor have their effect sizes been estimated in other samples. To identify QTL for DO in Japanese Black cattle, we conducted a genome-wide association study (GWAS) and estimated the effect of the QTL in the population.
The genetic parameters for calving interval-related traits have been evaluated until animals reach approximately 4 to 5 years of age in Japanese Black cattle (Wagyu Registry Association 2010). Thus, it is relative easy to obtain DO records from first to third or fourth parity. In this study, DO was evaluated as the average of three DO estimates (DO3) from primiparity to the fourth parity. The DO3 phenotypic data set included 15 488 records of reproductive females (Fig. S1). The data were filtered using six criteria to eliminate abnormal values and were corrected for the effects of farm and birth year (see legend of Fig. S1). The estimated direct heritability of DO3 was 0.03 using the numerator relationship matrix among 15 488 animals based on pedigree information (Fig. S1), consistent with previous reports for Japanese Black cattle (Oyama et al. 2002). The distribution of DO3 values was sufficiently wide to discriminate between lower and higher performance groups (Fig. S1). We selected 231 cows from the lower extreme (longer DO3; 85th percentile) of the distribution and 228 cows from the upper extreme (shorter DO3; 15th percentile). These samples were genotyped using the Illumina Bovine SNP50K BeadChip, comprising probes for 54 001 SNPs. A total of 33 303 autosomal SNPs that passed our quality control criteria (call rate > 99%, minor allele frequency > 0.01, Hardy–Weinberg equilibrium P > 0.001 and inclusion of the SNP on the Illumina Bovine HD BeadChip; Sasaki et al. 2013) were used for the association study. The association analysis was performed for 459 samples using gemma software based on a linear mixed model with genomic relationships (Zhou & Stephens 2012). The genomic inflation factor (λGC) in this analysis was 1.013, indicating no population stratification; therefore, the sampling was appropriate for an association study. A significant association (P < 0.05, Bonferroni-corrected) was detected for ARS-BFGL-NGS-115142 (97 368 169 bp, odds ratio = 1.703, P = 1.15 × 10–6) on bovine chromosome 2 (BTA2) (Fig. 1a, Table S1). This QTL has not previously been reported for reproductive traits in cattle (Fortes et al. 2013; Hu et al. 2013).
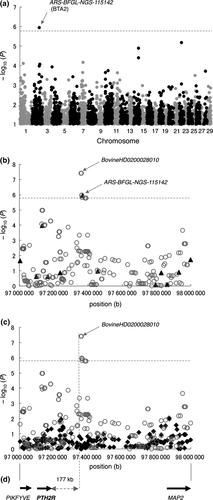
To characterise the region in more detail, the genotypes of 459 animals for 33 303 SNPs were imputed using beagle software (Browning & Browning 2009) with phased haplotype data inferred from 586 812 SNPs in a reference group of 1041 Japanese Black cattle (Sasaki et al. 2013). Six SNPs associated with DO3 were detected within the 8537-bp window spanning from bps 97 359 632–97 368 169 on BTA2 using gemma (P = 1.50 × 10−6–3.92 × 10−8, D′ = 1, r2 = 0.813–1) (Fig. 1b, Table S2). Except for the six SNPs on BTA2, we did not detect any SNPs that were significantly associated with DO3 (P < 0.05, Bonferroni-corrected). We then performed a conditional analysis to ascertain whether additional SNPs in the region demonstrated significant trait associations. Of the six SNPs, the BovineHD0200028010 genotype had the most significant association and was fitted in the mixed model as a covariate. After fitting, the associations of the other SNPs disappeared (Fig. 1c), indicating that the region contains a single QTL.
These SNPs were located on the telomeric side at a distance of 177 kb from the parathyroid hormone 2 receptor (PTH2R) gene (Fig. 1d, Table S3). Recently, a chromosomal conformation capture assay has revealed that distal loci become spatially close via DNA folding and act as long-range regulatory elements of gene expression (Schierding et al. 2014; Smemo et al. 2014). PTH2R is a receptor for the tuberoinfundibular peptide of 39 residues (TIP39), which regulates suckling-induced prolactin release in postpartum rats (Cservenak et al. 2010). In addition, TIP39-containing fibres in the thalamus terminate in the PTH2R neurons of arcuate nuclei (Dobolyi et al. 2012), which are a centre for pulsatile GnRH release (Wakabayashi et al. 2010). Thus, it is possible that the TIP39-PTH2R pathway, which could be modulated by binding between the associated region and the PTH2R regulatory region, is involved in the process of resuming ovarian cycles or ovulation during the postpartum period and in turn influences DO length.
To replicate the association and estimate the genetic variance explained by the SNP, we genotyped one of the most closely associated SNPs, BovineHD0200028010 (primer details are provided in Table S4) in 1778 animals that were randomly selected from the remainder of the cohort from the same farm used for the GWAS (Fig. S2). The Q allele was significantly associated with DO3 relative to the q allele (Tukey–Kramer post hoc test, P = 0.0245) (Table 1). The Q allele frequency was 0.632, indicating that it is common in Japanese Black cattle. The proportion of total genetic variance explained by the SNP was 1.72%. The effect of a Q-to-q allele substitution on DO3 was a decrease of 3.74 days (Table 1).
| No. of animals genotyped for the SNP | Q/q allele | Q allele frequency | Heritability | SNP effect on total genetic varianceb | A to G allele substitution effect_day (SE) | P-valuec |
|---|---|---|---|---|---|---|
| 1778d | G/A | 0.632 | 0.03 | 0.0172 | 3.74 (0.41) | 0.0245 |
- a Position is based on the UMD3.1 assembly of the bovine genome.
- b The effect of the SNP type was estimated as the least squares mean values of GLM analysis. The statistical model for the analysis included the farm, birth year and SNP type as fixed effects. The genetic variance explained by the SNP was calculated based on estimates of the SNP type effect and the frequency of the SNP (Falconer & Mackay 1996). Total genetic variance was estimated by the MTDF-REML programs. The effect size of a SNP was estimated as the proportion of the genetic variance explained by the SNP.
- c The result was tested by a one-way ANOVA, followed by the Tukey–Kramer test for multiple comparisons.
- d The mean and standard deviation for DO3 in 1778 animals was 103 ± 48 days.
In conclusion, the GWAS reported herein identified QTL associated with DO3 on BTA2 in Japanese Black cattle. The association was replicated in a sample of 1778 animals. The Q allele is common in the population, suggesting that it can be used to improve DO3 in Japanese Black cattle.
Acknowledgements
We would like to thank Emiko Watanabe and other laboratory members for generous support and valuable suggestions. This work was supported by the Ito Foundation (S.S.) and the Japan Racing and Livestock Promotion (Y.S.).




