Apolygus lucorum genome provides insights into omnivorousness and mesophyll feeding
Yang Liu, Hangwei Liu, Hengchao Wang, and Tianyu Huang contribute equally to this work.
Abstract
Apolygus lucorum (Miridae) is an omnivorous pest that occurs worldwide and is notorious for the serious damage it causes to various crops and substantial economic losses. Although some studies have examined the biological characteristics of the mirid bug, no reference genome is available in Miridae, limiting in-depth studies of this pest. Here, we present a chromosome-scale reference genome of A. lucorum, the first sequenced Miridae species. The assembled genome size was 1.02 Gb with a contig N50 of 785 kb. With Hi-C scaffolding, 1,016 Mb contig sequences were clustered, ordered and assembled into 17 large scaffolds with scaffold N50 length 68 Mb, each corresponding to a natural chromosome. Numerous transposable elements occur in this genome and contribute to the large genome size. Expansions of genes associated with omnivorousness and mesophyll feeding such as those related to digestion, chemosensory perception, and detoxification were observed in A. lucorum, suggesting that gene expansion contributed to its strong environmental adaptability and severe harm to crops. We clarified that a salivary enzyme polygalacturonase is unique in mirid bugs and has significantly expanded in A. lucorum, which may contribute to leaf damage from this pest. The reference genome of A. lucorum not only facilitates biological studies of Hemiptera as well as an understanding of the damage mechanism of mesophyll feeding, but also provides a basis on which to develop efficient control technologies for mirid bugs.
1 INTRODUCTION
Apolygus lucorum, a global Miridae pest, occurs throughout Asia, Europe, Africa and America, and causes large economic losses each year (Jiang et al., 2015; Lu & Wu, 2008, 2011b). This notorious pest has attracted renewed research attention due to its resurgence, and it is gradually increasing from a secondary to a major pest of cotton due to the drastically decreased usage of insecticides as a result of commercial adoption of Bt cotton (Lu et al., 2010; Wu et al., 2002). The area affected by mirid bugs and the associated yield losses in cotton both increased 3- to 5- fold from 1991 to 2010 (Lu & Wu, 2011b). To make things worse, A. lucorum not only damages cotton, but has also becomes a primary pest of other crops such as cereals, vegetables, and fruits, leading to more severe economic losses (Jiang et al., 2015; Lu & Wu, 2011b; Lu et al., 2010; Pan et al., 2015). In order to control A. lucorum, farmers increased the use of insecticides, which reduced or even counterbalanced the benefits of Bt cotton (Lu et al., 2010; Zhao et al., 2011). However, efficient environmentally friendly measures to control this pest are still lacking.
Apolygus lucorum possesses a variety of biological characteristics associated with successful insect pests, including strong environmental adaptability, high population growth rate, and strong dispersal ability (Lu & Wu, 2008, 2011b; Lu et al., 2007, 2009, 2010). The wide host range of A. lucorum is likely to be critical for its success, which increases the range of damaged crops and the difficulty of prevention. A. lucorum is omnivorous and is specifically phytozoophagous, and mainly phytophagous, feeding on over 200 plant species that comprise the majority of crops (Lu et al., 2012). As a polyphagous pest, A. lucorum grows and develops normally on different hosts, and it often switches food plants in the field depending on its highly developed chemosensory system (Pan et al., 2015), which has been found to considerably increase the population growth and survival of many phytophagous pests (Kennedy & Storer, 2000). In addition to feeding on plants, A. lucorum can also prey on some small insects or insect eggs, which further increases its environmental adaptation (Li et al., 2016; Lu et al., 2008; Yuan et al., 2013).
Due to its special mesophyll feeding pattern, A. lucorum causes severe damage to crops. Both nymphs and adults of A. lucorum can suck the sap of terminal meristems, tender leaves, flowers, bolls, and fruits, resulting in serious reduction in yields and quality of numerous crops (Jiang et al., 2015; Wu & Guo, 2005). A. lucorum is a typical mesophyll feeder, using the cytoplasm and nucleus of plant tissue as its food source (Sharma et al., 2014; Zhang et al., 2013). In the process of feeding, A. lucorum uses its stylets to pierce plant tissue and ingest the liquefied plant material, which is preliminarily degraded by enzymes in saliva the insect injects into the plant tissue (Lu & Wu, 2008; Wheeler, 2001; Zhang et al., 2015). This ‘lacerate and flush’ feeding action results in stunting, abscission of squares and bolls, and fruit malformation of plants. The key factor eliciting symptoms of the feeding damage is the saliva containing various digestive enzymes, rather than the mechanical damage (Sharma et al., 2014; Wheeler, 2001; Zhang et al., 2013), which might be the dominant reason for the economic importance of mirid pests (Frati et al., 2006).
Hemiptera is the most species-rich hemimetabolous order, including many notorious agricultural pests and human disease vectors (Cassis & Schuh, 2012; Lu et al., 2010; Wheeler, 2001). Several genome sequences have been reported, including from aphids (Li et al., 2019; Mathers et al., 2017; Richards et al., 2010; Wenger et al., 2017), plant hoppers (Wang et al., 2017; Xue et al., 2014; Zhu et al., 2017), kissing bug (Mesquita et al., 2015), seed bugs (Panfilio et al., 2019), bed bug (Benoit et al., 2016; Rosenfeld et al., 2016) and psyllids (Sloan et al., 2014), paving the way to uncovering the molecular and genetic mechanisms of many biological problems. Miridae (Hemiptera: Heteroptera: Cimicomorpha), also called plant bugs, is one of two most species-rich families in Hemiptera (with family Cicadellidae) and can be found in all major biogeographic regions of the world (Cassis & Schuh, 2012; Jung & Lee, 2012). With a wide range of food preferences and behaviours, some mirid bugs are pests of crops, whereas others are predatory natural enemies (Cassis & Schuh, 2012; Wheeler, 2001). Although studies have examined the feeding characteristics of many mirid bugs (Lu et al., 2007; Lu & Wu, 2011a), no reference genome data is available in Miridae, limiting in-depth studies of these pests. Here, we generated the first reference genome for a mirid bug, A. lucorum, to explore the genetic processes and molecular mechanisms of its omnivorousness and mesophyll feeding.
2 MATERIALS AND METHODS
2.1 Insect rearing and genomic sequencing
Apolygus lucorum nymphs and adults were collected from cotton fields at the Langfang Experimental Station of Chinese Academy of Agricultural Sciences, Hebei Province, China. A laboratory colony was established and maintained at 28 ± 1°C, relative humidity (RH) 60 ± 5%, and 14:10 light: dark (L:D) and reared on green beans and corn. An inbred strain of A. lucorum was developed by successive single-pair sib mating for 12 generations from this laboratory colony. This inbred strain was used for all genomic sequencing experiments in this study.
For Illumina sequencing, a short paired-end DNA library with a 400 bp insert size from a female adult A. lucorum was constructed using standard Illumina protocols, and sequenced on the illumina hiseq 2500 platform. For PacBio sequencing, two single-end libraries with ~20 kb insert size were constructed using PacBio SMRT from 100 female siblings of A. lucorum. PacBio long reads were sequenced using 18 SMRT Cells on the pacbio sequel system (Pacific Biosciences).
2.2 Assembly and polishing of contigs
A contig assembly was first carried out using canu (version 1.6; Koren et al., 2017) with parameters (ovsMethod = sequential genomeSize = 1g) based on PacBio long reads from pooled samples. This resulted in 18,403 contigs of total length ~1.97 Gb, contig N50 of 259 Kb.
Three rounds of contig polishing were performed. For the first round, contigs were polished using PacBio reads with the Arrow consensus caller in smrt-link version 5.1.0. The original bam files generated from PacBio Sequel were aligned with contig assembly by pbalign (version 0.3.1). Then, using arrow (version 2.2.2), we polished the assembly. For the second and third rounds, we used our in-house clean_adapter (-s 15 -a R1/2-adapter -r 75) and clean_lowqual (-e 0.001 -r 75) to filter out adaptors and low quality sequences in raw Illumina reads. After quality control, 140 Gb raw Illumina data from female A. lucorum generated 100 Gb clean data. The clean data were mapped to the contigs using bwa (version 0.7.12; Li & Durbin, 2009) and the assembly errors were corrected using pilon (version 1.22; Walker et al., 2014) with parameters (--fix bases --nonpf --threads 112 --minqual 20).
To filter haplotypic duplication in the contig assembly, we used purge_dups (version 1.2.3; Guan et al., 2020) with parameters (−2 -a 50) on the polished assembly. This resulted in a purged primary assembly of total length 1.03 Gb, contig N50 of 785 kb and a haplotig assembly of total length 936 Mb, contig N50 of 88 kb.
To assess the completeness of genome assembly, we run busco (version 3.0.2) using the insecta database (OrthoDB version 9), which contains 1,658 conserved insecta genes, with parameters ‘-m genome -sp pea_aphid -l insecta_odb9’. For the gene set completeness assessment, parameters was changed to ‘-m proteins -l insecta_odb9’.
2.3 Filtering contamination contigs
Because samples for PacBio sequencing were whole insects that may contain bacteria and parasites in the gut, and samples for Illumina sequencing were gut removed, we used clean Illumina data to filter possible contaminations in assembly. We used bwa (version 0.7.12) to align clean Illumina data with the assembly and if any contig had an Illumina coverage rate lower than 5%, it was removed. This resulted in a total of 80 contigs being removed from the genome. In order to identify further potential contaminations, we submitted the assembly to ncbi. After validation from contamination screening of NCBI, 22 additional contigs were reported as contaminated sequences from Lactococcus lactis, a common bacterium within the digestive tract of A. lucorum and were filtered.
2.4 Scaffolding with LACHESIS
About 5 g of fresh tissues from living insects were macerated and crosslinked using paraformaldehyde to capture the interacting DNA segments. Chromatin was subsequently digested with MboI (NEB), and biotinylated nucleotides were used to fill in the resulting sticky ends. Following ligation, a protease was used to remove the crosslinks. Finally, genomic DNA was extracted, sheared into 350 bp fragments using a focused ultrasonicator (Covaris), and fragments into which biotin had been incorporated were pulled down with streptavidin-coated magnetic beads. Purified DNA was then prepared and sequenced on an illumina hiseq instrument according to the manufacturer's recommendations.
After quality control, clean Hi-C paired-end reads were first mapped to the contig assembly by bowtie2 (version 2.3.4.3; Langmead & Salzberg, 2012), and then hic-pro (version 2.11.0; Servant et al., 2015) used the alignment to detect valid alignments and filter multiple hits and singletons. Finally, lachesis (Burton et al., 2013) was used to cluster, order and orient the contigs.
2.5 Transcriptome library preparation and sequencing
Total RNA (1 μg) was extracted by TRIzol reagent (Invitrogen) and was used to construct cDNA libraries. A total of 60 (20 samples × 3 replicates) individual unstranded cDNA libraries were prepared by ligating sequencing adaptors to cDNA fragments synthesized using random hexamer primers with NEBNext Ultra RNA Library Prep Kit for Illumina (NEB). The 20 samples included spawn, eight tissues (antenna, mouthpart, salivary gland, head, gut, leg, wing, body) from third instar nymphs, and 11 tissues (male antenna, female antenna, mouthpart, salivary gland, head, gut, leg, wing, male genital, female genital, body) from adults. Raw sequencing data were generated using an illumina hiseq 4000 system with the paired-end 150 bp (PE150) strategy. The average length of the insert sequence was 350 bp.
RNA samples from the whole body of A. lucorum in six different developmental stages including first to fifth nymph, and adult were also prepared for full-length transcriptome sequencing using the PacBio Iso-Seq protocol. Total RNA was extracted separately from the six samples, equal amounts of which were then mixed. The synthetic full-length cDNAs were selected to prepare three 20 kb SMRTbell template libraries for sequencing on a pacbio sequel instrument.
2.6 Genome annotation
Tandem repeats were identified by tandem repeats finder (version 4.07b; parameters: 2 7 7 80 10 50 2000 -f -m –d; Benson, 1999). Transposable elements (TEs) were identified using a combination of two methods: searching against the TE database (dfam 3.0, RepBase 20170127) by repeatmasker (version 4.0.9; with parameter: -nolow -no_is -norna -engine ncbi; http://repeatmasker.org) and searching against the TE protein database by repeatproteinmask (with parameter: -engine ncbi -noLowSimple); and constructing a de novo repeat library by repeatmodeler (version 1.0.8; parameters: -engine ncbi -database), followed by repeatmasker to find TE repeats. Because some repeat sequences in the repeatmodeler de novo library may come from simple repeats, low complexity regions, and duplicated protein-coding genes, we used two steps for filtering the non-TE repeat sequence to get a A. locurom specific TE library: first, the 1,062 repeat sequence of ‘unknown’ type were aligned to NR database by blastx (v2.7.1+) using 1e−5 as cutoff, and 14 of them were found to have homology with known non-TE protein-coding genes; second, the repeat sequence of ‘unknown’ type were subjected to sequence structure analysis, and 80 of them were found to have more than 20% regions covered by simple repeat or low-complexity sequences. In total, 94 ‘unknown’ repeat sequences in the repeatmodeler de novo library were filtered. After that, repeatmasker was used to find TEs based on the filtered de novo TE library.
The gene models in A. lucorum were predicted using augustus (version 3.3.2; Stanke et al., 2006) on the TE soft-masked genome, integrating evidence from RNA sequencing alignments, Isoform sequencing alignments and protein homology searches. For RNA-Seq, 20 paired-end datasets from different tissues were aligned with the genome using star (version 2.7.1a; Dobin et al., 2013). After filtering by filterBam in augustus, the sorted bam file was transferred to a hints file by bam2hints in augustus. Moreover, we used Iso-Seq to assist in gene prediction. gmap (version 2018-03–25) was used to align Iso-Seq sequences with the genome and blat2hints.pl in augustus was used to generate a hints file. Additionally, for protein homology evidence, all Hemiptera proteins in NCBI RefSeq were download. We aligned the proteins with the genome by tblastn (version 2.7.1+) using 1e−5 as cutoff and filtered those with less than 50% identity. We used exonerate (v2.2.0) to align the remaining proteins with the genome and used exonerate2hints.pl in augustus to generate a hints file. Finally, we combined all hints files from RNA-Seq, Iso-Seq and protein homology, and used augustus with the combined hints file to predict gene models, resulting in 23,106 gene models.
To get accurate gene sets, we filtered genes with less than 35 amino acids. We aligned protein sequences of gene models with the NR database in diamond (version 0.8.28) blastp using 1e−5 as a cutoff, and 16,187 gene models had homologous proteins in NR. We also aligned 20 RNA-Seq data sets with coding sequences of gene models in bwa (0.7.12) and 17,953 gene models had a coverage rate higher than 95%. We retained genes that either had homologous proteins in NR or had RNA support, resulting in 20,386 genes. After that, we detected 33 genes with two or more errors in start codons, stop codons or nontriplet length. We filtered those wrong genes and got the final official gene set (OGS) including 20,353 gene models.
For gene functional annotation, we aligned the protein sequences of genes with kegg, eggnog, nr, swiss-prot databases by diamond, an alternative replacement of blast, using 1e−5 as a cutoff and got the best hit. We also used interproscan (version 5.38-76.0) to search interpro databases to find motifs and domains. Taken together, 18,721 (91.98%) genes had homologous information in those databases, indicating that the OGS is reasonably accurate. Moreover, trnascan-se (versus 2.0) was used to find tRNAs with default parameters.
2.7 Evolutionary analysis
Nine sequenced hemipteran insects including Acyrthosiphon pisum, Myzus persicae, Bemisia tabaci, A. lucorum, Cimex lectularius, Rhodnius prolixus, Oncopeltus fasciatus, Laodelphax striatella, Nilaparvata lugens and Drosophlia melanogaster as an outgroup were used to infer gene orthology in OrthoFinder (Emms & Kelly, 2015) with default parameters. Phylogenetic tree and gene orthology results were displayed and annotated using Evolview (He et al., 2016). Expanded orthologous groups in A. lucorum were determined using a rank sum test compared to other eight insects in Hemiptera. Protein sequences of single copy genes from each species were aligned in muscle (version 3.8.1551; Edgar, 2004), then concatenated into one super-sequence. PhyML was used to reconstruct the phylogenetic tree based on the concatenated super-sequence with the LG + I + G + F model (Guindon & Gascuel, 2003). Divergence times among species were calculated in mcmctree (paml package, version 4.9; Yang, 2007). Calibration times were set according to a previous paper, minimum = 320 Ma and maximum = 390 Ma for D. melanogaster and A. lucorum (Misof et al., 2014). GO (Gene Ontology) annotation results were obtained from Interpro (Quevillon et al., 2005). GO enrichment analysis was performed using the OmicShare tools, a free online platform for data analysis (https://www.omicshare.com/tools/). The reciprocal BLAST best hit was used to calculate the synonymous mutation rate (Ks) by kaks_calculator 2.0 (Wang et al., 2010) with default parameters. Duplicate_gene_classifier in MCscanX (Wang et al., 2012) was implemented to classify the origins of the duplicated genes into different types.
2.8 Analysis of the digestive enzyme, chemosensory receptor, and detoxification enzyme genes
A set of described Hemiptera odorant receptors (ORs) and gustatory receptors (GRs) was used to search the A. lucorum gene sets by blastp with the cutoff e-value 1e−5. Multiple PSI-BLASTP searches were initiated with divergent ORs and GRs to find any additional annotated proteins that might belong to these families, and up to four iterations were used. Finally, some ORs and GRs were corrected manually. Ionotropic receptors (IRs), digestive enzymes, and detoxification enzymes were annotated using diamond (version 0.9.21.122) results compared to the nr database, uniprot database and kegg database with e-value 1e−5 and confirmed by InterProScan or eggNOG. To get a complete gene family set, reannotation of the gene family was performed. First, all digestive enzyme, chemosensory receptor, and detoxification enzyme genes got from former gene set was mapped to eight Hemiptera genomes by exonerate (version 2.2.0) with identity >35%, and exonerate2hints.pl was used to generate a hints file. Then, the region where these genes can map was used to predict gene models by augustus with the hints file, and short gene models (less than 200 bp) were filtered. The predicted gene model that doesn't exist in former gene set was added to the gene family sets. The protein sequences were aligned using muscle (version 3.8.1551; Edgar, 2004). mega x (Knyaz et al., 2018) was used to reconstruct the phylogenetic tree for digestive enzymes, chemosensory receptors, and glutathione S-transferases (GSTs) with the neighbour-joining method. PhyML was used to reconstruct the phylogenetic tree for cytochrome P450 monooxygenases (P450s). To analyse gene expression, clean reads of each sample were mapped to A. lucorum gene sets using bowtie2 (version 2.2.5; Langmead & Salzberg, 2012), and then rsem (version 1.2.12; Li & Dewey, 2011) was used to count the number of mapped reads and estimate the TPM (transcripts per million) value of each gene. The phylogenetic tree and heatmap were generated by itol (Letunic & Bork, 2016).
3 RESULTS
3.1 Chromosome-level genome assembly and recent expansion of DNA and LINE TEs
Due to the high heterozygosity of wild Apolygus lucorum, we used female adults from a 12-generation inbred strain for genomic sequencing. First, we produced 140 Gb paired-end 150 Illumina reads, and estimated the genome size of A. lucorum to be 1.03 Gb based on k-mer analysis (Figure S1). Then, we produced 103 Gb (~100× coverage) single molecule real-time (SMRT) sequences with an N50 read length of 14 kb on the PacBio Sequel platform (Table S1), and generated an assembly comprising 3,818 contigs with a total length of 1.02 Gb and contig N50 length of 785 kb (Tables 1, S2, S3). Finally, with Hi-C scaffolding, 1,016 Mb (99%) contig sequences were clustered, ordered and assembled into 17 large scaffolds with scaffold N50 length 68 Mb (Figure 1a, S2 and Table S4), the longest of which was 117 Mb and the shortest 16 Mb. To validate this assembly, we mapped paired-end clean Illumina reads onto it, resulting in a mapping rate of 99.39%. Furthermore, the assembled genome size was comparable to the estimated genome size. Based on ORTHODB version 9 of Insecta, 1,610 (97.1%) busco genes were found in the genome, including 1,564 (94.3%) complete and 46 (2.8%) fragmented busco genes. Moreover, 75 (4.5%) ‘duplicate’ busco genes were found in A. lucorum, comparable or lower than most compared species, suggesting that the current reference assembly contains few redundant fragments derived from highly heterozygous genomic regions (Figure 1b). Apart from the assembled nuclear genome, we also assembled the mitochondrial genome that has 99.7% identity with the published mitochondrion (Wang et al., 2014; Figure S3). In total, read mapping, genome size estimation, busco assessment and assembly of the mitochondrion demonstrated that our genome assembly is reasonably accurate, and the majority of the genome has been successfully assembled.
| Number | Size (bp) | |
|---|---|---|
| Assembly feature | ||
| Assembled contigs | 3,818 | 1,023,255,709 |
| Contigs N50 | 785,215 | |
| Anchored scaffolds | 17 | 1,015,881,696 |
| Scaffolds N50 | 68,132,828 | |
| Estimated genome size (by k-mer analysis) | 1,030,903,443 | |
| Genome annotation | ||
| Tandem repeats | 468,796 | 72,964,816 |
| Transposable elements | 2,273,150 | 655,921,134 |
| Gene models | 20,353 | 27,622,289 |
Note
- Each scaffold represents a natural chromosome.
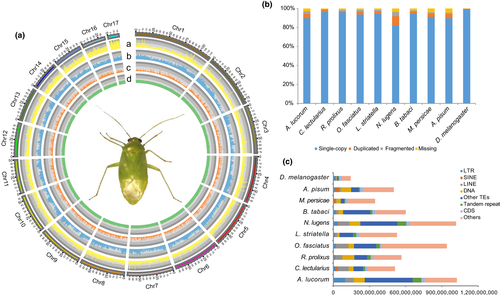
A total of 20,353 annotated gene models were regarded as the official gene set (OGS), with average CDS length of 1.4 kb and average exon number of 6.6, which are comparable to other published hemipteran insects (Table S5; Figure S4). Furthermore, based on orthodb version 9 of Insecta, 1,617 (97.5%) busco genes were found in OGS (Figure S5), including 1,579 (95.2%) complete and 38 (2.3%) fragmented busco genes, which was consistent with the busco assessment result for the genome assembly (Simao et al., 2015; Figure 1b). This indicated that our gene prediction pipeline has successfully obtained nearly all potential genes in the assembled genome (Table S6). In the OGS, 18,721 (91.98%) genes have homology evidence in at least one database among kegg, nr, eggnog, interpro and swiss-prot (Figure S6). Additionally, a total of 427 tRNAs for all 20 essential amino acids and suppressor are detected (Table S7).
The highly continuous genome enables us to study TEs comprehensively. In total, 656 Mb (64.10%) TEs were annotated in the A. lucorum genome, which is much more than the closest relatives C. lectularius (218 Mb) and N. lugens (489 Mb) with similar genome sizes (Figure 1c and Table S5, S8). Moreover, the genome size of A. lucorum is also the largest of compared species, indicating that TEs extensively contribute to the genome size. In A. lucorum, LTR (98 Mb), LINE (73 Mb) and DNA (88 Mb) elements are the major types of TEs, and LTR is considerably in excess of that from other compared insects (Figure 1c). Considering that most other genomes are poorly continuous, the comparison of TE content here may be biased. The top three families of LTR are Gypsy (59 Mb), Pao (16 Mb) and Copia (9 Mb; Figure S7). In addition, we found that DNA and LINE transposons had a recent activity burst at 3% and 4% divergence rates, respectively (Figure S8). This could be essential in promoting adaptive evolution for A. lucorum, as the recent explosion of active TEs can bring more genetic variations for adaptive selection and are often related to the gene enrichment of functions such as stress response and adaptations (Liu et al., 2018).
3.2 Gene expansion and recent gene burst promote environmental adaptability
To gain insights into an evolutionary perspective for A. lucorum, a whole genome-based phylogenetic analysis was performed with nine hemipteran insects A. pisum, M. persicae, B. tabaci, A. lucorum, C. lectularius, Rhodnius prolixus, O. fasciatus, L. striatella, N. lugens, and D. melanogaster as the outgroup (Table S9). All protein-coding genes from these ten species were used to construct gene orthologous groups, resulting in 13,933 orthologous groups. The phylogeny showed that A. lucorum diverged from C. lectularius about 168 million years ago (Ma) and from A. pisum about 275 MYA (Figure 2a).
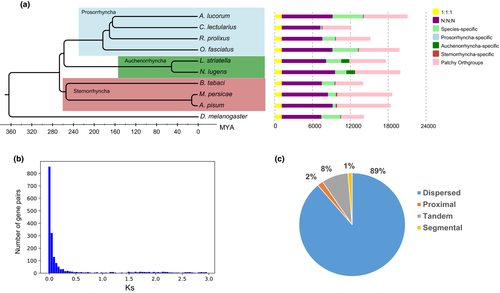
Compared to the other eight hemipteran species, 1,086 expanded orthologous groups (rank sum test) containing 2,872 genes were identified in A. lucorum. Gene ontology analyses observed significant enriched GO terms (Fisher's exact test, p-value < .01) involved in odorant recognition, including sensory perception of smell (GO: 0007608) and sensory perception of chemical stimulus (GO: 0007606; Figure S9), which provided clues for the extremely broad host plant ranges of A. lucorum, as host plant selection mainly relies on odorant recognition for many herbivorous species. In addition, enriched GO terms associated with digestion in A. lucorum were also observed, such as hydrolase activity, acting on glycosyl bonds (GO: 001698), hydrolase activity, hydrolysing O-glycosyl compounds (GO: 0004553) and polygalacturonase (PG) activity (GO: 0004650). Particularly, PG is an essential enzyme for digestion, which hydrolyses pectin substances and then destroys plant cell walls (Markovič & Janeček, 2001). In summary, these expanded genes could play an important role in the severe damage on a wide range of plants, as PGs can hydrolyse the pectin substances and then destroy the plant cell walls and ORs could promote the pest search for host.
Novel genes were mostly generated from gene duplication, which is recognized as a driving force of evolution. Using a within-genome reciprocal best blast hit, 2,609 paralogue pairs were identified, and distribution of synonymous distances (Ks values) showed that 1,502 (58%) paralogue pairs had a Ks value smaller than 0.3, suggesting that most gene duplications possibly occurred in a recent period (Figure 2b). Among all the recent duplicated pairs, 89%, 8%, and 2% were classified as dispersed duplication, tandem duplication, and proximal duplication, respectively, indicating that recent duplicated genes in A. lucorum are mostly derived from small local scale gene duplications, instead of whole genome duplication (Figure 2c). Notably, 642 (42%) recent duplicated paralogue pairs are included in 675 (62%) expanded orthologous groups, so recent gene duplication events have made considerable contribution to the gene expansion in A. lucorum.
3.3 Expansion of digestive enzyme genes promotes processing of diverse foods
Our genome-wide analysis showed that A. lucorum has a comprehensive digestive enzyme spectrum including 55 PGs, 49 carboxypeptidases, 39 cathepsins, seven alpha-amylases, 224 serine proteases (SPs), 44 aminopeptidases, 48 phospholipases, 73 lipases, and 12 glucosidases (Table S10). Compared with the other eight hemipteran insects, A. lucorum had a comprehensive digestive enzyme spectrum, with a unique group of PGs and a significantly expanded group of SPs (Figure 3a).
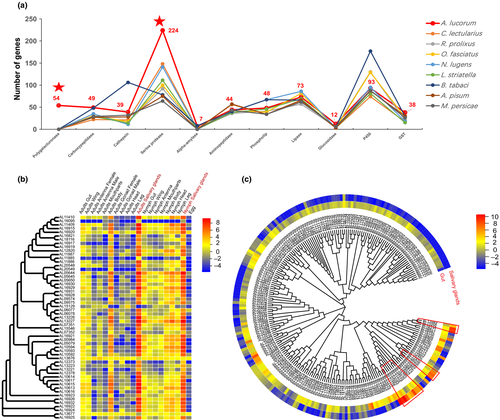
PG is a group of plant cell wall-degrading enzyme, ubiquitous in fungi, bacteria, and plants. It is also found in Hemiptera and Coleoptera, predicted to be horizontally transferred from fungi (Wybouw et al., 2016; Xu et al., 2019). PG activity has been detected in the salivary glands of Hemiptera including some aphid species, but is especially common in mirid bugs (Zhang et al., 2015). Most notably, 55 unique PGs were detected in A. lucorum, but that were not found in the other eight Hemiptera including two aphids. Many PGs were arranged in tandem on the genome with high identity, which suggests recent replication (Figure S10). The duplication of PGs in A. lucorum might contribute to expanding the host plant range of this insect (Wybouw et al., 2016). The expression profile showed that 55 PGs were specifically expressed in salivary gland with high expression levels, indicating that the salivary gland of A. lucorum has a very high ability to synthesize PGs (Figure 3b). A large amount of multi-form PGs are secreted by salivary glands and injected into the plant to destroy the tissues, elicit feeding-damage symptoms and induce much larger lesions than other piercing-sucking insects (Wheeler, 2001).
SPs are involved in various physiological processes of insects, such as digestion, development and innate immunity (Di Cera, 2009). The digestive SPs are essential in degrading proteins into free amino acids and inactivating toxic compounds from food sources. The expansion of SPs is the most obvious among the digestive enzyme genes of A. lucorum, and a total of 224 SPs were identified in the genome, the most of all nine Hemiptera species (Figure 3a), the phylogenetic tree of SPs within four Prosorrhyncha species exhibited most subclades are presented in A. lucorum (Figure S11). The type and abundance of digestive enzymes is related to the nature of the food source that an insect can assimilate (Agusti & Cohen, 2000). Carnivorous insects should secrete greater amounts of proteases than herbivorous insects (Zeng & Cohen, 2000). Some strictly phytophagous mirid bugs even lack detectable digestive proteases (Cohen & Wheeler, 1998). The expansion of SPs in A. lucorum can improve its digestive capacity and may contribute to its omnivorous feeding habit, mainly phytophagous with prey to complement (Li, et al., 2019; Xu et al., 2019). The slight expansion of SPs was also observed in bed bug, which is consistent with its carnivorous feeding habit. Transcriptome analysis showed that 201 (90%) SPs were specifically expressed in the gut, in contrast, the other 21 SPs that clustered in three clades (Figure 3c) were highly expressed in the salivary gland and the expression levels were much higher than the SPs expressed in the gut, indicating that these 21 SPs may function in digestion and degrading toxic compounds from the plants in the mesophyll feeding process of A. lucorum.
3.4 Rapid evolution of chemosensory receptors expands the range of host plants
Insects have developed a highly efficient chemosensory recognition system to recognize and distinguish chemical signals that are essential to host plant selection (Blomquist & Vogt, 2003). Reception of chemical signals in insects is mediated by three families of chemosensory receptors, including ORs, GRs, and IRs. The size of chemosensory receptor families is largely related to the complexity of chemical signals the insect detects. A large number of chemosensory receptors containing 135 ORs, 57 GRs and 33 IRs were identified in the A. lucorum genome, indicating that this insect has a sensitive and specific perception system to distinguish complicated chemical signals in the environment (Figure 4a).
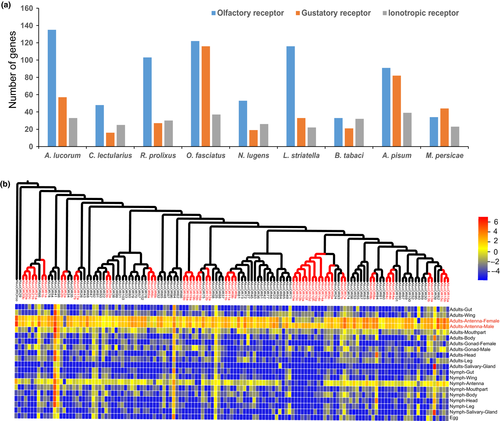
OR is the first chemosensory receptor gene family identified in insects, which locates in the dendritic membrane of odorant receptor neurons and function as a heterodimer with a highly conserved noncanonical OR coreceptor (Orco). The number of ORs varies considerably in different insects, from ten in the body louse to up to 400 in the ants, and protein sequence identities of ORs are also widely divergent, suggesting ORs are minimally conserved genes in insects (Kirkness et al., 2010; Zhou et al., 2012). We identified 135 ORs in A. lucorum, the most among the nine Hemiptera species. ORs expanded three-fold in A. lucorum compared with the closely related species C. lectularius. The phylogenetic analysis that showed 40% of the OR genes (55) were contained in several clades with high protein sequence identity (>80%), indicating that OR has experienced recent gene replication (Figure 4b). Transcriptome analysis showed that 99% of ORs were highly expressed in antenna, as antenna is the major olfactory perception organ in insects. The expansion of ORs, mostly due to the recent replication in A. lucorum, may be related to its extensive hosts and host-switching behaviour.
GRs, another kind of chemosensory receptor gene, are expressed in gustatory receptor neurons in taste organs (Clyne et al., 2000). Unlike ORs, which mainly sense volatile compounds, these GRs sense nonvolatile compounds that the insects directly contact with gustatory sensilla, including carbon dioxide (CO2), sugars, bitter compounds, and salts (Vosshall & Stocker, 2007). Similar to ORs, the number of GRs also greatly varies in different insects, and 237 GRs were reported in Spodoptera litura from Lepidoptera (Cheng et al., 2017). In contrast, only 10 GRs were found in the honey bee Apis mellifera from Hymenoptera (Robertson & Wanner, 2006), associated with their different feeding habits. Our results show that the number of GRs varies greatly in different Hemiptera, from 21 in L. striatella and N. lugens to 116 in O. fasciatus. GRs showed significant expansion in A. lucorum compared to the two related carnivorous species C. lectularius (16) and R. prolixus (27), indicating that phytophagous insects have more GRs than carnivorous insects, which is consistent with the adaptation of phytophagous insects to multiple hosts. Expression profile analysis showed that GRs exhibit different expression patterns and most GRs are expressed in various tissues (Figure S12), which is consistent with the previous reports in C. lectularius and O. fasciatus, indicating the diversity of GR functions.
IRs, which evolved from the ionotropic glutamate receptor superfamily (iGluRs), were recently identified as involved in chemosensory reception (Benton et al., 2007). Different from ORs or GRs, IRs exhibit diverse functions and participate in olfactory, gustatory and auditory sensation. The sequences of IRs are more conserved than OR and GR, and the numbers of IRs are also more stable in different insect species. We identified 33 IRs in A. lucorum, comparable to the other eight Hemiptera species. Expression profile studies showed that 88% of IRs are expressed in antennae, suggesting that most IRs have olfactory functions. However, some IRs were highly expressed in tissues other than antennae, reflecting the diversity of IR functions (Figure S13).
3.5 Expansion of detoxification enzymes contributes to degrading toxin
Agricultural pests usually employ an efficient detoxification system containing various enzymes to overcome numerous toxins in food sources or the environment. We performed a genome-wide analysis of the two important detoxification enzymes GST and P450 to study the mechanism of detoxification in A. lucorum.
GST is a superfamily of multifunctional isoenzymes involved in the cellular detoxification of various physiological and xenobiotic substances, which is highly related to insecticide resistance in insects, as it can directly detoxify the insecticides (Fang, 2012). A total of 38 GSTs were identified in A. lucorum, which is the highest of the nine Hemiptera species (Figure 5a, Table S11). Expression profile analysis showed that most GST genes were expressed in various tissues, in line with their detoxification function (Figure S14). The number of GSTs was more than twice as high as the other three closely related true bug species C. lectularius (17), R. prolixus (20), and O. fasciatus (27), indicating a significant expansion in A. lucorum. Phylogenetic analysis of the GSTs of four true bugs showed three A. lucorum-specific branches, which contained 58% GST genes (22; Figure 5b), suggesting the GSTs experienced a recent species-specific expansion in A. lucorum, enabling better detoxification of toxic substances and adaptation to the environment.
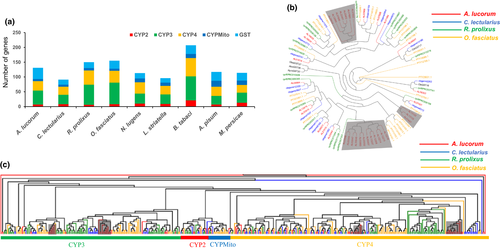
P450s constitute the largest and most functionally diverse class of insect detoxification enzymes, including four distinct clades CYP2, CYP3, CYP4 and CYPMito (Li et al., 2007). A total of 93 P450s containing seven CYP2, 47 CYP3, 32 CYP4, and seven CYPMito have been identified in the genome of A. lucorum. Most P450s also showed similar expression patterns as GSTs and were simultaneously expressed in multiple tissues (Table S11, Figure S15). Members of the CYP3 clade have been implicated in the oxidative detoxification of plant secondary metabolites and synthetic insecticides. CYP3s were significantly expanded in A. lucorum compared with the closely related species C. lectularius, but were comparable to the gene numbers of other two true bugs R. prolixus and O. fasciatus, which have different feeding habits. The phylogenetic tree of P450s exhibited four A.lucorum-specific branches (Figure 5c), suggesting that P450s experienced similar species-specific expansion as GSTs, enhancing the detoxification activity in A. lucorum.
4 DISCUSSION
Although long and ultra-long reads from the third-generation sequencing technologies have been widely adopted, large animal genomes with high heterozygosity still pose great challenges for genome assembly. Here, we present a 1.02 Gb chromosome-scale reference genome of Apolygus lucorum accomplished using PacBio and Hi-C data. Due to the limitations in available sequencing and assembly technologies, the current genome assembly is still far from perfect. Future sequencing that can decrease the required amount of DNA to achieve single individual sequencing, and improvements in the assembly algorithms on resolving heterozygosity problem, will contribute to improving assembly quality. With the genome sequence of A. lucorum, we are able to comprehensively characterize the TEs and find a recent explosion of DNA and LINE TEs. In addition, we identified a wave of recent gene duplications, which may be responsible for the damage capacity of A. lucorum. As one of a few species in Hemiptera with published genomes and the first sequenced mirid bug species, A. lucorum has the potential to become a model species of Miridae, and the reference genome of A. lucorum not only facilitates biological studies of Hemiptera, but also contributes to in-depth studies of damage mechanisms of agricultural pests.
The A. lucorum genome can be used to uncover the molecular and genetic mechanism of specific biological problems in Miridae. At the genomic level, we clarified that PG is unique in mirid bugs, which is the reason for the serious damage of A. lucorum caused by its mesophyll feeding; moreover, these PG genes are also specific targets for the control of this important pest. Differences in the living environments and eating habits exhibited a great influence on the abundance of related genes, as many genes associated to digestion, chemosensory perception, and detoxification expanded in A. lucorum compared to other Hemiptera species. Consistent with its phytozoophagy, the numbers of plant-feeding associated genes identified in A. lucorum are more similar to distantly related phytophagous species than the two closely related carnivorous species. In contrast, the numbers of the carnivorous-related genes in A. lucorum are similar to those of carnivorous species, which reflects the environmental plasticity of the insect genome. As the number of sequenced genomes in Miridae increases, the environmental adaption and damage mechanism will be further clarified in the future.
In recent years, with the worldwide expansion of Bt cotton planting area, the damage caused by mirid bugs gradually increased (Wu et al., 2002). Various ecological and biological strategies have been used to control A. lucorum, with some successes (Lu & Wu, 2011a; Lu, et al., 2007). But these management strategies have not reached the optimal effect, so the control of A. lucorum still relies on the application of synthetic insecticides (Lu & Wu, 2011b; Zhen et al., 2018). Large-scale and long-term use of insecticides results in several ecological and environmental problems, especially increased resistance to insecticides, and A. lucorum was reported to have different levels of resistance to many insecticides (Zhen et al., 2018). There is an urgent need for developing new strategies to control this pest, and the genome data provide us abundant gene targets associated with specific biological processes in A. lucorum. By targeting these key genes, RNA interference (RNAi), genetic regulation, behavioural regulation and other environmentally friendly strategies can be introduced to control A. lucorum. Therefore, the genome and transcriptome data as well as the candidate drug targets provide a basis to develop efficient control technologies for mirid bug pests, protecting global agricultural safety.
ACKNOWLEDGEMENTS
This work was funded by National Natural Science Foundation of China (31621064 to K.W., and 31672095 to Y.L.), National Key R&D Program of China (2017YFD0200400 to YL, and 2016YFC1200600 to W.F.), Agricultural Science and Technology Innovation Program & The Elite Young Scientists Program of CAAS, Fundamental Research Funds for Central Non-profit Scientific Institution (No. Y2017JC01), the Agricultural Science and Technology Innovation Program Cooperation and Innovation Mission (CAAS-XTCX2016), and the Fund of Key Laboratory of Shenzhen (ZDSYS20141118170111640). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
AUTHOR CONTRIBUTIONS
K.W., W.F., and G.W. designed and led the research. Yang. L., B.Y., Bi.L., S.Z., X.Z., B.W., and Yan. L. prepared DNA and RNA for sequencing. H.W., Bo.L., Y.Z., F.J., Y.R., S.W., and W.F. assembled the genome and generated the gene set. Yang. L., H.L., T.H., and B.Y. annotated the gene families. Yang. L. and H.L. analysed the transcriptome. Yang. L., H.L., H.W., G.W., W.F., and K.W. wrote the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
All the raw sequencing data and genome data in this study have been deposited at NCBI as a BioProject under accession PRJNA526332. Genomic sequence reads have been deposited in the SRA database with BioSample: SAMN11095929. Transcriptome sequence reads have been deposited in the SRA database with BioSample: SAMN13219540. Genome assembly and gene annotation has been deposited at DDBJ/ENA/GenBank under the whole genome shotgun project accession WIXP00000000. The version described in this paper is version WIXP02000000. The genome assemblies and annotation files are also available at the website ftp://ftp.agis.org.cn/~fanwei/Apolygus_lucorum_Genome/.