Genomic analysis and behavioral ecology records of the vulnerable Kong skate (Okamejei kenojei)
Graphical Abstract
Wild populations of cartilaginous fish (sharks, skates, rays, and chimaeras) are encountering challenges. Here, we are unveiling genomic data and behavioral ecological records of Okamejei kenojei, a species listed in the IUCN Red List of Threatened Species, aiming to offer insights into the conservation and environmental adaptability of cartilaginous fish.
Cartilaginous fish (sharks, skates, rays, and chimaeras) are an important group of vertebrates facing the risk of regional extinction due to factors such as overfishing (Pacoureau et al. 2021; Finucci et al. 2024; Worm et al. 2024). However, knowledge of their reproduction and conservation remains limited. Understanding the genetic diversity, population structure, and reproductive behaviors of cartilaginous fish can offer valuable insights into their reproductive strategies and ecological adaptability, which are crucial for the conservation of these vulnerable species. Here, we present a chromosome-level draft genome as well as whole genome resequencing data of seven other individuals for the Kong skate (Okamejei kenojei), a species that has been reported to have a population decrease based on fisheries resource surveys (Deng & Jin 2001; Rigby et al. 2021). Our analysis revealed lineage-specific genomic changes in O. kenojei, including the expansion of 69 gene families. We also assessed their genetic diversity, which showed a decline in population size over the past million years. Furthermore, we examined the egg-laying behavior of 24 adult female specimens and the incubation period of 35 embryos, indicating a low reproductive capacity. In conclusion, our findings suggest that the genetic diversity of the current wild population of O. kenojei is relatively healthy, but their reproductive output is low. Without proper management and protection, this could lead to irreversible impacts on the population, highlighting the importance of conservation efforts for this species.
Cartilaginous fishes (sharks, skates, rays, and chimaeras), as the sister group to the osteichthyans, provide important insights into the evolution of vertebrates (Gillis et al. 2022; Marlétaz et al. 2023), while also playing crucial roles in maintaining the stability of marine ecosystems. However, due to factors such as fishing, these organisms are experiencing rapid declines. Their slow life histories, long life spans, and low reproductive capability make it difficult for wild populations to recover (Finucci et al. 2024; Worm et al. 2024). The Kong skate (O. kenojei), also known as the swarthy skate or ocellate spot skate (www.marinespecies.org) and the senior synonym of Raja porosa (Song et al. 2023), was once one of the most economically valuable fish in the northern coastal areas of China. These fish, characterized by their flat bodies, a habit of dwelling on the seabed, and a diet consisting of shrimp and other benthic organisms, play an important role in sustaining the ecological balance of their habitats. However, due to overfishing and environmental changes, along with a variety of other contributing factors, the wild populations have dramatically diminished (Rigby et al. 2021), leading to its recognition as a vulnerable species on The IUCN Red List of Threatened Species™ (www.iucnredlist.org). These challenges highlight the urgency of conservation. Therefore, assessing the survival status and reproductive capacity of O. kenojei and exploring effective conservation strategies become one of the most hopeful methods to save these creatures.
In this study, we assembled a chromosome-level draft genome of O. kenojei, which had a genome size of 2.71 Gb (Fig. 1a; Figs S1,S2, Supporting Information). The assembly included 1954 contigs anchored into 50 chromosomes, with chromosome lengths ranging from 2.53 to 194.74 Mb (Fig. 1a) and a contig N50 of 3.8 Mb (Table S1, Supporting Information). The assembly's BUSCO score was 94.30% (Tables S2,S3, Supporting Information) and it exhibited a strong synteny with the little skate (Leucoraja erinacea) (Fig. S3, Supporting Information). Repetitive elements accounting for 66.90% of the O. kenojei genome were identified, including DNA transposons (4.08%), long interspersed nuclear elements (LINEs, 37.15%), long terminal repeats (LTRs, 12.98%), short interspersed nuclear elements (SINEs, 2.52%), and tandem repeats (TRF, 5.20%) (Fig. 1a). We identified a total of 17 793 gene families in O. kenojei, including 4584 multi-copy (more than two copies) and 7658 single-copy gene families. The phylogenetic tree was based on the single-copy homologous genes of eight species, revealing that O. kenojei was most closely related to L. erinacea and Amblyraja radiata, with a divergence time around 63.66 million years ago (Fig. 1a,b). In O. kenojei, 69 gene families had undergone specific expansion, while 10 gene families had undergone contraction (Fig. 1b). These changes might be related to the ecological adaptability of O. kenojei in coastal environments (Fig. S4, Supporting Information).
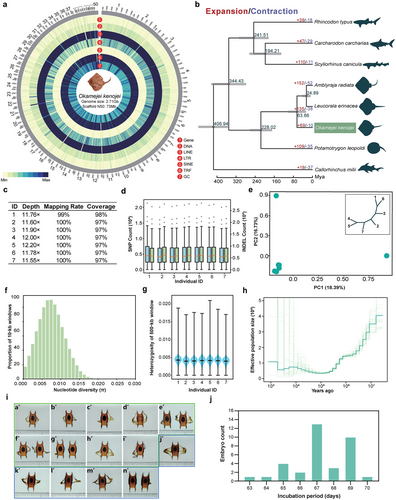
We further conducted whole-genome resequencing on seven skate individuals. Resequencing was achieved at an average 11.83× depth with 97% average genome coverage (Fig. 1c). A total of 73 641 903 single nucleotide polymorphisms (SNPs) were identified, which were located on 50 assembled chromosomes of O. kenojei. 398 757 SNPs were in exonic regions, 25 369 233 SNPs were in intronic regions, and 48 000 460 SNPs were in intergenic regions (Fig. 1d; Table S4, Supporting Information). Following the filtering of InDel data from O. kenojei populations, 11 789 713 InDel loci were obtained; 4 922 339 were insertions and 6 867 374 were deletions (Table S4, Supporting Information). Principal components analysis (PCA) outcomes aligned with the neighbor-joining tree, Kin-ship analysis, and population structure, indicating that these individuals constitute a single group without population structure (Fig. 1e; Figs S5,S6, Supporting Information). The resequencing results showed that the average nucleotide diversity (θπ) of the skate population was 0.0075 and their genome-wide heterozygosity ranged from 0.0039 to 0.0042 (Fig. 1f,g). The proportion of runs of homozygosity in the O. kenojei genome was relatively small (about 0.74%, Fig. S7, Supporting Information). These metrics indicated relatively healthy genetic diversity, reflecting a large historical population size. Nevertheless, multiple sequentially Markovian coalescence analysis suggested a general declining trend in the population of O. kenojei over the past one million years (Fig. 1h).
In addition, a morphological analysis was conducted on 103 eggs, including 11 eggs without yolks (non-yolk eggs), produced by 24 female skates under captive conditions (Dataset S1, Supporting Information). Results showed that the average rate of egg production (including non-yolk eggs) by this group of females during the egg-laying season was 2.06 egg cases per day. Data of egg case production from two female skates, bred separately and labeled A and B, showed their production rates were 0.8 and 0.4 egg cases per day, respectively (Fig. 1i; Dataset S2, Supporting Information). The difference between the overall average and individual egg-laying rates suggested that there might be certain high-laying females within the group, as well as the potential impact of group behavior on egg-laying rates. The morphological images indicated that eggs produced by the same individual shared more morphological characteristics (Fig. 1i; Datasets S1,S2 Supporting Information). They could be easily distinguished by certain morphological characteristics (such as the anterior apron, Fig. S8, Supporting Information). This suggested that the morphological features of the eggs might have a potential genetic basis. In addition, to assess the potential impact of temperature on the development of skate embryos, the study examined the hatching behavior of 35 embryos. It was found that, in an environment with an average temperature of 19.8°C (Tables S5,S6, Supporting Information), the embryos required 63–70 days to hatch (Fig. 1j; Tables S5,S6 and Video S1, Supporting Information), with the growing degree days (GDDs) ranging from 1247.4 to 1386. These results indicated that the reproductive output of O. kenojei was limited.
Research on the reproduction of cartilaginous fish, including O. kenojei, is important but challenging due to difficulties in data collection, environmental variability, and individual differences. These challenges could lead to an underestimation of extinction risks for these species. Here, due to the limited sample size (n = 7) and the focused sampling areas, accurately estimating the population size across nearly 10 000 years poses a challenge with the current dataset (Fig. 1h). There could be discrepancies between our estimates and the actual population dynamics. Furthermore, a previous study on mitochondrial DNA sequences highlighted the complexity of the O. kenojei populations (Misawa et al. 2019). Therefore, further investigations in more regions and with more samples are essential for a comprehensive understanding. Additionally, our findings offer valuable insights into the behavioral ecology of O. kenojei, emphasizing that more attention should be focused on the conservation and management of O. kenojei. Notably, the observed GDDs (at about 19.8°C) for O. kenojei differ from those previously reported for other cartilaginous fish (Onimaru et al. 2018; Gillis et al. 2022; Vazquez et al. 2022), suggesting that habitat or other environmental factors may influence these variations. However, the variability in incubation periods even within the same environmental conditions indicates a complex interplay of genetic and environmental factors affecting embryonic development. Further research, with an expanded ecology dataset, is essential to evaluate GDDs in cartilaginous fish (Gao et al. 2022). Additionally, considering the impacts of global climate change, assessing the thermal stress on embryonic development in cartilaginous fish becomes increasingly important (Di Santo 2019; Wheeler et al. 2021). Therefore, understanding these thermal response mechanisms is also crucial for the conservation of marine species that are vulnerable to climate impacts. In conclusion, our study suggests that despite the historically large population sizes of O. kenojei, conservation efforts and long-term population monitoring are still necessary due to factors such as their limited reproductive output and the potential impact of environmental variables on their reproductive cycle.
ACKNOWLEDGMENTS
We thank Ms. Xinling Yu and Mr. Yanteng Liu for collecting skate samples. We thank all members of Workshop 0 of Laizhou Mingbo Aquatic CO., LTD., for the assistance in breeding skates and collecting eggs, and we are grateful to Mr. Cheng Li for helping with the transportation of the skates. This work was supported by the National Key R&D Program of China (2022YFC3400300).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JBFCOM000000000. The version described in this paper is version JBFCOM010000000 and the project number for the resequencing data is PRJNA1110068.