RNA Sequencing, De novo assembly, functional annotation and SSR analysis of the endangered diving beetle Cybister chinensis (= Cybister japonicus) using the Illumina platform
Abstract
Cybister chinensis Motschulsky, 1854 (synonym Cybister japonicus Sharp, 1873) is a beetle found in ponds and irrigation canals near rice fields regulating the aquatic faunal community through predation. However, due to loss of natural habitats, use of pesticides, and invasion of alien species the beetle is threatened. With lack of understanding at the trophic ecology and genomics level, the conservation study is hindered to a larger extent. In the present study, Illumina HiSeq 4000 platform has been used to unravel the whole-larval transcriptome of the beetle. A total of 20,129 non-redundant unigenes were assembled from 67,260,666 clean read sequences. About 18,743 unigenes found a homologous match in any one of the databases like PANM, UniGene, Swiss-Prot, Clusters of Orthologous Groups (COG), Gene Ontology (GO), KEGG, and InterProScan. While the zinc finger domains topped the unigene hits, about 660 enzymes (2695 sequences) participating in metabolism, environmental information processing, genetic information processing and organismal system pathways were recorded. Furthermore, the HSP70 class, Toll-like receptors 4, insulin-receptor substrate, and AMP activated protein kinase showed conspicuous presence in the larval transcriptome. Out of a total of 12,491 unigene sequences examined, 1968 SSRs were detected. Majority of them were dinucleotide repeats with six iterations followed by trinucleotide and tetranucleotide repeats with five and four iterations, respectively. This is the first report of cDNA resources from C. japonicus till date. The data would be crucial for the assessment of the beetle in the wild and making an inventory for utilisation in future genomics and ecological studies.
Introduction
Cybister japonicus Sharp (Coleoptera: Dytiscidae), commonly known as the diving beetle, is native to Southern Asia. The beetle species is located in paddy fields, spending their larval phase in water and migrating to other habitats when adult. The species is predatory in habit feeding on aquatic insects, excepting for the 3rd instar larvae that feed on vertebrate animals such as tadpoles (Ohba and Inatani 2012). This is important for understanding trophic ecology of the species under the insect conservation program (Ohba 2009a). The predatory ability of the beetle against Japanese Encephalitis Vector, Culex tritaeniorhynchus and the larval populations of Anopheles sinensis has been reported (Ree 2005; Ohba and Takagi 2010). Further, morphological and ultrastructural insights to the antennae and labial palp of the beetle has generated sufficient information on the chemoreceptors, so critical for detecting food (Song et al. 2017) and chemical communication between male and female sexes (Song et al. 2016).
The population of C. japonicus that was once seen aplenty in the rice fields and ponds, has seen a dramatic disappearance in the mainlands. The 2010 Tokyo Red List released by the Tokyo metropolitan government categorized C. japonicus as an extinct species. The species of the beetle has been threatened due to loss of natural habitats, invasion of alien species, use of pesticides in the paddy fields, and limitations in their food resources (Ohba and Inatani 2012). Considering the economic value of the species as biological control agents and the imposed endangered status, the species has been categorized under the protected list. Some recent efforts have provided insights to the feeding preferences of the species as one of the strategies towards the insect conservation plans (Ohba 2009a, 2009b). Having said that, the genetic background of the species is still unexplored making it difficult to extract phenotypic cues and provide aggressive survival strategies of the insect in the natural habitat. The National Centre for Biotechnology Information (NCBI) Taxonomy browser for C. japonicus contain the details of odorant-binding protein 1 & 2 (Song et al. 2016), cytochrome-oxidase subunit-I & II, histone III, and wingless proteins (https://www.ncbi.nlm.nih.gov/protein/?term = txid398594[Organism:noexp]) (Miller et al. 2007).
Next-generation sequencing (NGS) platforms have been increasingly used to map the genetic regulatory circuits and provide insights to survival and adaptation strategies of commercial and threatened species of insects (Morozova and Marra 2008; Wheat 2010; Patnaik et al. 2016). Moreover, the NGS platforms have been useful to generate transcriptome data and analyse the complexity of the transcriptome in non-model species of insects (Oppenheim et al. 2015; Patnaik et al. 2015; Patnaik et al. 2016) revealing the candidate genes involved in stress responses, chemosensory processes, metabolism, and immune processes of insects including the beetles (Vongsangnak et al. 2016; Duan et al. 2017; Wei et al. 2017). The Roche 454 FLX Titanium platform-based pyrosequencing technology provided large-scale gene discovery in the coleopteran pest, Eucryptorrhynchus chinensis (Liu and Wen 2016), ground beetles, Carabus iwawakianus and Carabus uenoi (Fujimaki et al. 2014), and the banana weevil, Cosmopolites sordidus (Valencia et al. 2016). The Illumina second-generation sequencing technology has taken the lead in the de novo transcriptome analysis of the coleopteran insect, Dastarcus helophoroides (Zhang et al. 2014), four species of luminescent beetles (Wang et al. 2017) and many other species utilized in the integrated pest management schemes. In this study, we used Illumina HiSeq 4000 and de novo assembly to analyse the transcriptome of the endangered diving beetle, C. japonicus. Further, the TransDecoder program was used to shortlist the putative transcripts with open-reading frames (ORF) and cluster the same to non-redundant unigenes. Functional annotation of the unigenes was conducted using the COG, GO, Interpro, and KEGG databases. We identified the simple sequence repeats (SSRs), that once validated could be used to understand the species variability and polymorphism in the populations. The genetic resource cataloguing of C. japonicus would assist in understanding the genera diversity and may be one of the tools required for insect conservation programs.
Materials and Methods
Sample collection, processing, and RNA extraction
C. japonicus is a protected species under the Endangered Wildlife law. Presently, it is categorized as the species of Least Concern (LC) in the Red List. Hence, no permit was required for the collection of the species. For RNA extraction, samples of C. japonicus adults were collected from Deogcheon-ri, Gujwa-eup, Jeju-si, Jeju-do, Korea. The whole-body of the adults were ground to fine powder in liquid nitrogen using mortar and pestle. The total RNAs were isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer's recommendations and stored at −80°C till further use. The total RNA was treated with RNase-free DNase I (Qiagen, Hilden, Germany) as described in the manufacturer's protocol. The integrity of the DNase-treated RNA was evaluated by using the NanoDrop 2000 spectrophotometer (NanoDrop, Wilmington, DE, USA) and gel electrophoresis. Before Illumina sequencing, the RNA samples were observed for RIN (RNA integrated number) > 7 using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
cDNA library construction and Illumina Sequencing
The cDNA library construction and Illumina HiSeq 2500 sequencing was conducted at GnC Bio-Company Limited, Daejeon, Korea. Briefly, the total RNA was processed for mRNA purification using magnetic beads with oligo (dT). Using a fragmentation buffer, the mRNA was sheared into shorter fragments at 94°C for 5 min. Then, first-strand cDNA was synthesized from the mRNA fragments using N6 random primers and reverse transcriptase. The second-strand cDNA was synthesized using buffer, dNTPs, DNA polymerase I, and RNase H. The synthesized double-strand cDNA went through an end-repairing process using an End Repair (ERP) mix. Further, a single ‘A’ overhang was added to the 3′-end of the end-repaired fragments. This would prevent the fragments to ligate to one another during the adapter ligation process. Sequencing adapters were added to the ends of the cDNA and analysed by agarose gel electrophoresis. PCR amplification enriched the cDNA libraries and a paired-end transcriptome sequencing was conducted on Illumina HiSeq 4000 platform. This generated 2 × 100 base pairs (bp) read length. The raw data obtained from Illumina sequencing has been submitted to Sequence Read Archive (SRA) at National Centre for Biotechnology Information (NCBI) with accession number SRR5182815. The C. japonicus transcriptome is held under the BioProject PRJNA362249 and BioSample SAMN06236838. The datasets and the assembled contigs information are available for download from http://bioinfo.sch.ac.kr/submission/.
Assembly and annotation
After the generation of raw reads, the assembly was assessed using FASTQC (Version 0.11.3) (www.bioinformatics.babraham.ac.uk/projects/fastqc/). The command-line tool Cutadapt (http://code.google.com/p/cutadapt/) was used as default parameters (for paired-read ends: aADAPT1-Aadapt2; −o out1. fastq –p out2. fastq in1. fastq in2. fastq) for filtering adapter-only sequences (number of nucleotides of recognized adapter ≤13, and number of nucleotides excluding the adapter ≤35). Next, the low-quality reads were trimmed using the base-calling program Phred (quality scores ≥20) (Ewing & Green 1998). Finally, the possible GC-content bias was removed to get quality reads for an accurate de novo assembly. Trinity RNA-Seq assembly (release v2.0.4); http://gthub.com/trinityrnaseq/trinityrnaseq/) was used for the de novo assembly of the quality reads. For Trinity assembly, a K-mer of 25 and a minimum allowed length of 25 nucleotides was allowed. Trinity, combines clean reads to longer contigs (sequences contain overlap without gaps). The TransDecoder program (https://transdecoder.github.io/) was implemented to extract only the protein-coding genes from assembled contig sequences. The default parameters of a minimum length of 100 amino acids and a log-likelihood score of 0 were used for the identification. TIGR Gene Indices Clustering Tools (TGICL) program (Pertea et al. 2003) was used for clustering and defining unigenes (sequences without Ns and which could not be extended on either ends).
The assembled unigenes were annotated using blastx (BLAST, the basic local alignment tool) and an E-value of <1e-5 to several protein databases such as Protostome DB (PANM-DB), Swiss-Prot, COG, GO, Interpro, and KEGG (Kanehisa et al. 2012; Mitchell et al. 2015). The NCBI nucleotide database, UniGene DB was also used for the annotation (blastn; E-value of <1e-5) of the assembled unigenes. PANM-DB was also utilized for the homology mapping of the assembled unigenes with reference to E-value, identity, similarity distribution, and the hit and non-hit ratio. Further, the database was also used to decipher the top-hit species distribution for the assembled unigenes of C. japonicus. PANM-DB (http://malacol.or.kr/blast/PANM.html) is an efficient resource that have been developed to annotate the molluscan, arthropod, and nematode assembled sequences when compared with the NCBI non-redundant database.
Functional analytics of unigenes using COG, Blast2Go, KEGG, and InterPro domains
The unigenes were compared to the protein sequences available in the Cluster of Orthologous Groups (COG) library using blastx and then mapped to the COG classification (Tatusov et al. 2003). Further, the blastx results were imported to the Blast2Go pipeline (Conesa et al. 2005) for protein domain analysis, GO terms, and KEGG annotation. For COG classification, the unigenes were distributed under 25 different classes. The unigenes were also annotated to GO terms under biological process, cellular component, and molecular function categories (Ashburner et al. 2000). Blast2Go pipeline was also used to predict the conserved domains in the unigenes using the Interpro Scan function. To analyse unigene-relevant biochemical pathways, KEGG Orthology (KO) classification was used. The annotated unigenes under the KO classification were distributed to ‘Environmental Information Processing’, ‘Genetic Information Processing’, ‘Metabolism’, and ‘Organismal Systems’ categories.
Identification of SSRs
For the identification of microsatellites (especially SSRs) in the functional unigenes, MISA (MicroSAtellite identification tool) program v1.0 software (http://pgrc.ipk-gatersleben.de/misa/ accessed September, 2016) was used (Thiel et al., 2003). SSRs from 2 (dinucleotides) to 6 (hexanucleotides) were analysed with the repeat motif types. SSRs of 1 repeat (mononucleotides) was not consider due to a possibility of getting homopolymer sequences in Illumina platform.
Results and Discussion
De novo assembly of C. japonicus transcriptome
The Illumina HiSeq 4000 paired-end sequencing was performed on a cDNA library constructed from C. japonicus adults. A total of 675.45 million raw reads (67.54 GB) with a total of 33,772,889 read pairs (10,199,412,478 bp) were processed. Adapter trimming and the removal of contaminating and low-quality sequences identified 9,500,253,195 bp of filtered reads (Table S1). This accounts for 6.9% of raw reads discarded and an average length of trimmed reads of 140.6 bp. After quality control measurement, 99.58% (67.26 GB) high-quality reads were obtained with an average and N50 length of 139.4 bp and 151 bp, respectively. The high-quality reads accounted for a total of 174,853 contigs with the largest contig having a size of 26,683 bp. Almost 38.59% of contigs were ≥500 bp. The average length, N50 length, and GC% of contigs were 773.6 bp, 1337 bp, and 36.34%, respectively. Of the total contigs obtained, 82,133 sequences were predicted as protein-coding using the TransDecoder program. The mean length and N50 length improved to 1454.7 bp and 2520 bp, respectively, while the GC% was 38.35%. After the analysis, a total of 65.88% of sequences were having lengths ≥500 bp. Clustering of the putative protein-coding genes resulted in 20,129 sequences (37,631,641 bases) called unigenes having average and N50 lengths of 1869.5 bp and 2738 bp, respectively. The unigenes ranged in length from 140 bp to 26,683 bp. The exhaustive summary of the de novo assembly, TransDecoder analysis, and clustering has been given in Table 1.
| Sequencing | |
|---|---|
| Raw reads | |
| - Number of sequences | 67,545,778 |
| - Number of bases | 10,199,412,478 |
| Clean reads | |
| - Number of sequences | 67,260,666 |
| - Number of bases | 9,374,578,390 |
| - Average length of contig (bp) | 139.4 |
| - N50 length of contig (bp) | 151 |
| - GC % of contig | 41.91 |
| High-quality reads (%) | 99.58 (sequences), 91.91 (bases) |
| Contig information | |
| - Total number of contig | 174,853 |
| - Number of bases | 135,271,568 |
| - Mean length of contig (bp) | 773.6 |
| - N50 length of contig (bp) | 1,337 |
| - GC % of contig | 36.34 |
| - Largest contig (bp) | 26,683 |
| - No. of large contigs (≥500 bp) | 67,473 |
| After TransDecoder analysis | |
| - Total number of sequence | 82,133 |
| - Number of bases | 119,475,586 |
| - Mean length of sequence (bp) | 1,454.7 |
| - N50 length of sequence (bp) | 2,520 |
| - GC % of sequence | 38.35 |
| - Largest sequence (bp) | 26,683 |
| - No. of large sequences (≥500 bp) | 54,112 |
| Unigene information | |
| - Total number of unigenes | 20,129 |
| - Number of bases | 37,631,641 |
| - Mean length of unigene (bp) | 1,869.5 |
| - N50 length of unigene (bp) | 2,738 |
| - GC % of unigene | 37.98 |
| - Length ranges (bp) | 140–26,683 |
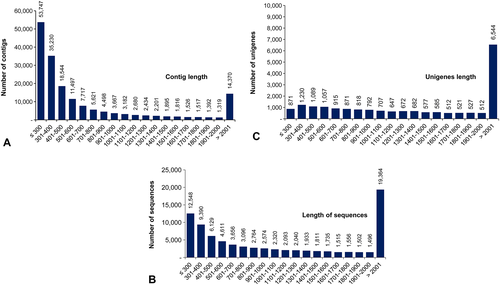
Further, we analysed the assembled contigs, TransDecoder sequences, and unigenes based on the distribution of sizes (Fig. 1). In case of the contig lengths, maximum sequences were ≤500 bp (107,521 sequences out of 174,853). The number of contigs gradually reduced with an increase in contig lengths till 2000 bp. Only 14,370 contigs (~8.22% sequences) were having sizes of >2001 bp (Fig. 1A). Only 34.17% of sequences ≤500 bp were found to be putatively protein-coding. A total of 19,634 sequences (23.58% of the total protein-coding sequences) were of length > 2001 bp (Fig. 1B). Further, 6544 unigenes (32.51%) out of a total of 20,129 sequences were of lengths >2001 bp. Only 15.85% of unigenes were having lengths ≤500 bp (Fig. 1C).
Sequence annotation of C. japonicus unigenes
For the annotation of unigenes, the sequences were queried against public databases (both protein and nucleotide) using blastx (BLAST, the basic local alignment tool) at an E value <1e-5. The locally curated database, PANM-DB version 2.0; October 2016 release (http://malacol.or.kr/blast/PANM.html) was preferred over the NCBInr database. Using PANM-DB the transcriptome processing across the phylum Arthropoda, Nematoda, and Mollusca could be conducted with high efficiency and greater speed and accuracy as compared with the NCBInr database (Kang et al. 2016b). PANM-DB was successful in annotating 18,656 transcripts out of a total annotation of 18,743 transcripts. This annotation efficiency was high when compared with all the other databases. Out of the sequences annotated in PANM-DB, 65.8% were having length ≥ 1000 bp. In total, 15,414 (76.58%), 15,662 (77.81%), 12,317 (61.19%), and 11,151 (55.4%), unigenes were annotated to the Swiss-Prot, COG, GO, and InterProScan databases, respectively (Table 2). A total of 1133 unigenes annotated to KEGG database suggesting the association of the sequences to functional pathways. Further, 38.66% of transcripts annotated to the UniGene database of nucleotide sequences. To summarize, annotation hits for 18,743 sequences out of a total of 20,129 C. japonicus unigenes were found. Out of 18,743 all transcript hits, 65.59%, 31.05%, and 3.36% of the transcripts were having lengths ≥1000 bp, 300–1000 bp, and ≤300 bp, respectively.
| Databases | All transcripts | ≤300 bp | 300–1000 bp | ≥1000 bp |
|---|---|---|---|---|
| PANM-DB | 18,656 | 621 | 5,760 | 12,275 |
| UniGene | 7,782 | 160 | 1,362 | 6,260 |
| ‘PPP'Swiss-Prot | 15,414 | 371 | 3,950 | 11,093 |
| COG | 15,662 | 376 | 4,092 | 11,194 |
| GO | 12,317 | 277 | 2,914 | 9,126 |
| KEGG | 1,133 | 9 | 158 | 966 |
| InterProScan | 11,151 | 30 | 1,757 | 9,364 |
| ALL | 18,743 | 629 | 5,820 | 12,294 |
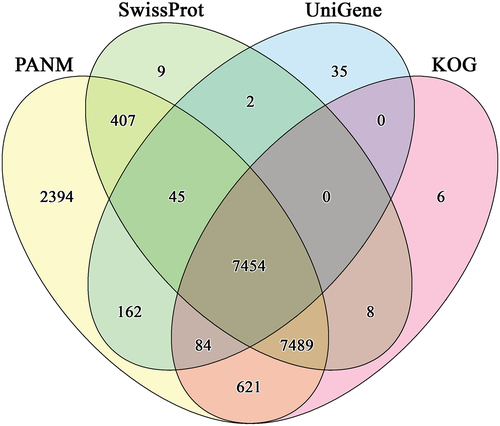
A Venn diagram for the shared and unique unigenes of C. japonicus according to PANM, Swiss-Prot, UniGene, and COG databases is shown in Figure 2. The maximum number (2394 sequences) of unique transcript annotation was observed with PANM-DB as compared to 35, 9, and 6 sequences for UniGene, Swiss-Prot, and COG databases, respectively. A total of 7454 transcripts were annotated in all the four databases. Further, the three protein databases shared 7489 unigenes. The sequence annotation result confirms the utility of PANM-DB as a potent resource for annotation hits for specific species like the arthropod, C. japonicus. The database has been efficiently utilized in the previous studies to characterize the de novo assembled unigenes of molluscs and arthropods alike (Park et al. 2016; Seong et al. 2016; Kang et al. 2016a). While all the earlier reports utilized version 1.0 of the database (Kang et al. 2015), the present study utilized the latest version released with updated sequences (Kang et al. 2016b). In fact, the latest release shows nearly two times more number of sequences (7,571,246) compared with the previous version (4,051,323 sequences).
Homology matching of C. japonicus unigenes
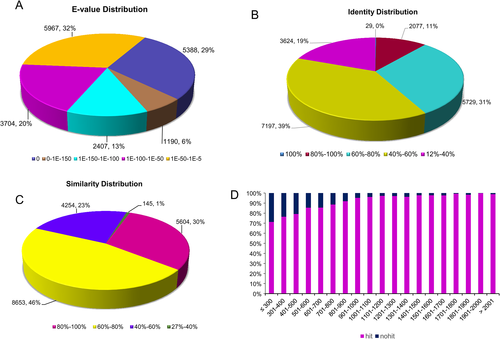
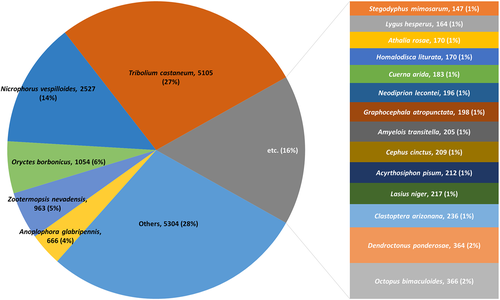
The E-value, identity distribution, similarity distribution, and the number of unigene hits and non-hits for C. japonicus unigenes were matched with the homologous protein sequences in PANM-DB (Fig. 3). A greater proportion of the unigenes showed an E-value distribution of 1E-50 – 1E-5 (32%), followed by an E-value of 0 (29%), 1E-100 – 1E-50 (20%), and 1E-150 – 1E-100 (13%) (Fig. 3A). In the identity distribution analysis, most of the unigenes showed an identity of 40–80% with homologous sequences in PANM-DB. About 39% and 31% of unigenes showed an identity of 40–60% and 60–80%, respectively (Fig. 3B). The annotation of C. japonicus unigenes to the proteins in the database showed that about 46% unigenes are 60–80% similar (Fig. 3C). The number of unigene non-hits decreased considerably with an increase in the unigene lengths. This suggests that with larger unigene lengths there is a greater possibility of obtaining sequence conservation through domains (Fig. 3D). Further, among the PANM-DB annotated unigenes, 27% of the homologous species showed the best match (blast results) to the coleopteran beetle, Tribolium castaneum (Fig. 4). Another 14% of unigenes showed matches with the burying beetle, Nicrophorus vespilloides. Among the best matched beetle, the T. castaneum genome sequence is available with the assembly encoding 16,500 genes (Tribolium genome consortium, 2007). Further, sufficient information on the expressed sequence tags (ESTs) and cDNA transcriptomes of the beetle have been reported that has improved the genome understanding (Park et al. 2008; Morris et al. 2009; Altincicek et al. 2013). Similarly, the transcriptome resource of N. vespilloides has been sequenced using Illumina platform with identification of genes leading to antimicrobial immunity (Palmer et al. 2016).
Functional annotations of C. japonicus unigenes
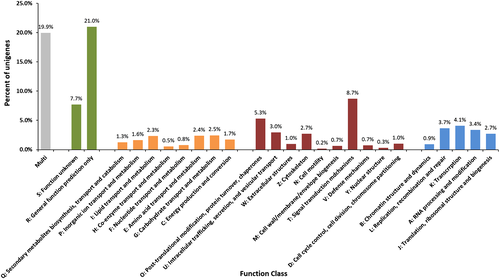
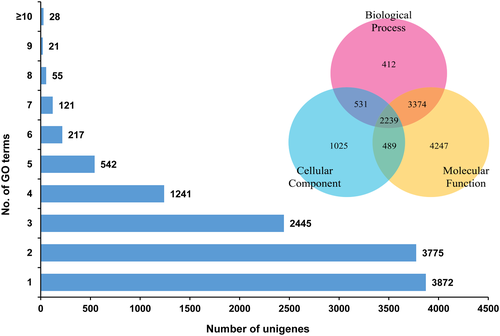
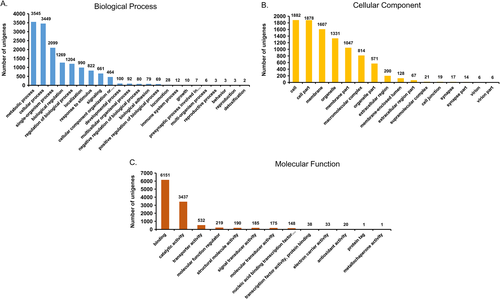
We annotated the C. japonicus unigenes based on the COG classification. COG classifies the unigenes to 25 diverse functional categories. For the C. japonicus unigenes, the top categories of classification includes general function prediction (21% of unigenes), signal transduction mechanisms (8.7%), function unknown (7.7%), post-translational modifications, biosynthesis, transport and catabolism (5.3%) (Fig. 5). About 19.9% of C. japonicus unigenes were annotated under the multi-category. The least represented COG categories were cell motility (0.2%), nuclear structure (0.3%), co-enzyme transport and metabolism (0.5%), cell wall/membrane/envelope biogenesis (0.7%), and defense mechanisms (0.7%). GO functional predictions for the 12,317 GO annotated unigenes showed a maximum of 4247 sequences classified under the ‘molecular function’ category, followed by 1025 under ‘cellular component’, and 412 under ‘biological process’ category (Fig. 6). A three-way Venn diagram also predicts 3374 unigenes shared between ‘biological processes’ and ‘molecular function’ categories. A total of 2239 unigenes shared between all the three GO functional terms. Only 531 and 489 unigenes were shared between ‘biological process’ and ‘cellular component’ and ‘cellular component’ and ‘molecular function’ category, respectively. About 31.44% of C. japonicus unigenes annotated to a single GO term closely followed by 30.65% unigenes annotated to two GO terms. Under ‘biological process’ category, the unigenes were predominantly classified to metabolic process (3545 unigenes), cellular process (3449 unigenes) and single-organism process (2099 unigenes) (Fig. 7A) while under the ‘cellular component’ category most unigenes were assigned to cell (1882 unigenes), cell part (1878 unigenes), and membrane (1607 unigenes) sub-categories (Fig. 7B). The predominant sub-categories under the ‘molecular function’ category included binding (6151 unigenes) and catalytic activity (3437 unigenes) (Fig. 7C). Classification of unigenes to GO term categories is only suggestive of predicted function and in no way an actual representation of function. The GO term annotations are associated with evidence codes (EC) and most of these (over 95%) are computationally-derived sources such as ‘inferred from electronic annotations (IEC)’, ‘inferred from sequence or structural similarity’, and inferred from reviewed computational analysis (RCA)’ (Rhee et al. 2008). The EC distribution of C. japonicus unigenes also suggest that over 98% of sequence annotations to GO terms were inferred from electronic annotations (Data not shown). EC describes the type of experimental support that links the unigenes to the GO ontologies. Hence, EC such as ‘inferred from direct assay (IDA)’ and ‘inferred from genetic interaction’ provides a more superlative evidence for the gene products annotated to the GO molecular function, cellular component, and biological process ontologies compared with IEC (Hill et al. 2008).
To identify the active biological pathways in C. japonicus, we mapped the unigene sequences to the reference canonical pathways in the KEGG database (Table S2). A total of 2695 sequences were assigned to 115 KEGG pathways belonging to the (Table S3) metabolism, organismal systems, environmental information processing, and genetic information processing categories. Almost 90% of sequences classified to metabolism pathway, most predominantly nucleotide metabolism (731 sequences), metabolism of cofactors and vitamins (437 sequences), and carbohydrate metabolism (294 sequences). Among nucleotide metabolism, purine metabolism and within metabolism of cofactors and vitamins, thiamine metabolism was the largely populated KEGG pathways. The metabolism of the terpenoids polyketides contained a lesser number of sequences under the secondary metabolites category. These pathways may be required to establish the ecological dependence, interactions, and evolutionary relationships (Pankewitz and Hilker 2008). Two sequences classified to ubiquinone and other terpenoid-quinone biosynthesis pathway consistent to the detection of methylhydroquinone, and toluquinone 2, 3-dimethylquinone in the larval transcriptome of the carabid beetle, Chlaenius cordicollis (Holliday et al. 2015). Among the organismal system category, the sequences were classified to the immune system pathways (194 sequences; 98 sequences of which classified to T-cell receptor signalling pathway). Included within the 2695 sequences annotated to KEGG Orthology category are 660 sequences with Enzyme Commission (EC) numbers out of which 143 belonged to carbohydrate metabolism. Further, Interpro domain analysis also provided a significant understanding of the putative functions of the unigenes (Table 3). Most conspicuously represented domains represented within the sequences are the zinc-finger C2H2 type, protein kinase type, and leucine-rich repeat type domains. These repeats are ubiquitous in most regulatory proteins with metabolic or immune functions. As understood, zinc-fingers are small but repeated units of protein motifs assisting in protein–protein contacts (Gamsjaeger et al. 2007). C2H2-type zinc fingers are the most common DNA-binding motifs found in eukaryotic transcription factors and has ability to bind to RNA and protein targets (Brayer and Segal 2008). Such motifs have been screened in the global transcriptome of many arthropods including pine-tip moth, Rhyacionia leptotubula (Zhu et al. 2013), cotton boll weevil, Anthomonas grandis (Firmino et al. 2013), and the Colorado potato beetle, Leptinotarsa decemlineata (Kumar et al. 2014). Protein kinase type domains are characteristic features of enzyme kinases that mediate many immune and metabolic signalling pathways in intracellular milieu. The protein kinase domain along with the zinc finger C2H2-type and RNA recognition motif domain has been noticed in the salivary gland transcriptome of potato leafhopper, Empoasca fabae (DeLay et al. 2012). The leucine-rich repeat region is prominently noticed in pattern-recognition receptors that is a molecular signature for the recognition of microbes, such as the Toll receptors (Akira et al. 2006). Overall, the C. japonicus transcripts belonged to putative proteins having interaction motifs participating in immune and metabolic signalling pathways.
| Domain | Description | unigenes |
|---|---|---|
| IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 566 |
| IPR007087 | Zinc finger, C2H2 | 421 |
| IPR015880 | Zinc finger, C2H2-like | 390 |
| IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain | 362 |
| IPR011009 | Protein kinase-like domain | 339 |
| IPR000719 | Protein kinase domain | 284 |
| IPR016024 | Armadillo-type fold | 269 |
| IPR011993 | PH domain-like | 251 |
| IPR013083 | Zinc finger, RING/FYVE/PHD-type | 251 |
| IPR015943 | WD40/YVTN repeat-like-containing domain | 236 |
| IPR012677 | Nucleotide-binding alpha-beta plait domain | 225 |
| IPR032675 | Leucine-rich repeat domain, L domain-like | 217 |
| IPR017986 | WD40-repeat-containing domain | 216 |
| IPR017441 | Protein kinase, ATP binding site | 209 |
| IPR011989 | Armadillo-like helical | 208 |
| IPR000504 | RNA recognition motif domain | 200 |
| IPR001680 | WD40 repeat | 186 |
| IPR008271 | Serine/threonine-protein kinase, active site | 185 |
| IPR016040 | NAD(P)-binding domain | 167 |
| IPR001611 | Leucine-rich repeat | 159 |
Identification of SSRs
The Illumina 4000 based transcriptomics data provided an excellent resource for identification of SSR markers in the C. japonicus transcripts. SSR markers in the cDNA sequences have been used for gene polymorphism and population genetic studies. As these markers are transferable across species, and are obtained at a greater speed than conventional approaches (including the hybrid capture method, loci selection from available genetic information, and loci transferable from closely related species), these act as potent resource for molecular ecologists and conservation biologists (Karaiskou et al. 2008; Uliano-Silva et al. 2014). Out of the total of 20,129 unigene sequences for C. japonicus, 12,491 sequences were analysed for SSR identification. We screened 1968 SSRs from 1349 of these sequences which were classified from dinucleotides to hexanucleotides with 2 to 6 repeats units, respectively. A total of 343 sequences were found to have more than one SSR. As a matter of caution and to avoid mis-representation, we avoided using the single-nucleotide repeats that may have been generated due to Illumina-platform homopolymer generation. The dinucleotide repeats were the maximum, followed by tri- and tetranucleotide repeats. All the information regarding the screened SSRs from C. japonicus unigenes have been provided in Table S4. Using the BatchPrimer 3.0 (You et al. 2008), we were able to elucidate the primer pairs flanking the SSR motifs under the default parameters such as primer lengths of 18–23 nucleotides, PCR product size of 100–300 bases, Tm of 50–70°C and primer GC content of 30–70%.
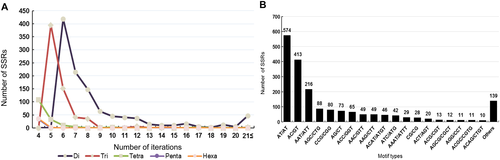
Further, as shown in Figure 8A, a maximum of 419 dinucleotide repeats showed six iterations, followed by 214 and 148 repeats in seven and eight iterations, respectively. The trinucleotide repeats were found more in five iterations while the tetra-, penta-, and hexanucleotide repeats were found in four iterations. Among the repeat motif types, AT/AT types (574 repeats) were more predominant followed by AC/GT (413 repeats) among the dinucleotide repeats. Among the trinucleotide repeat motifs, AAT/ATT was the most predominant with 216 repeats (Fig. 8B).
Conclusions
This is the first exhaustive survey of transcriptomics resources from the threatened beetle, C. japonicus that was once used in the insect conservation plans. We utilized the Illumina 4000 sequencing platform to decipher the transcriptome reads, applied de novo assembly method and TransDecoder program to identify the putative protein-coding genes and annotated the same against public databases for the functional classification and identification of adaptation-related genes. The transcripts were accorded functional categories and an important group of transcripts were identified that are basic to adaptation phenotypes in the species. We have also screened SSR markers from the unigenes that would be potent in identification of species diversity.
Acknowledgments
This work was supported by the grant entitled “The Genetic and Genomic evaluation of Indigenous Biological Resources” funded by the National Institute of Biological Resources (NIBR201503202), “Analysis of genetic characteristics of endangered species” funded by the National Research Foundation (NRF-2017R1D1A3B06034971) and Soonchunhyang University Research Fund.




