Typing of Panton-Valentine leukocidin-encoding phages carried by methicillin-susceptible and methicillin-resistant Staphylococcus aureus from Italy
Abstract
Panton-Valentine leukocidin (PVL) is the hallmark of community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) but can also be found in methicillin-susceptible S. aureus (MSSA) sharing pathogenic and epidemiological characteristics of CA-MRSA. PVL is encoded by two co-transcribed genes that are carried by different staphylococcal bacteriophages. We applied an extended PCR-based typing scheme for the identification of two morphological groups (elongated-head group and icosahedral-head group I phages) and specific PVL phage types in S. aureus isolates recovered in Italy. We examined 48 PVL-positive isolates (25 MSSA and 23 MRSA) collected from different hospital laboratories from April 2005 to May 2011. spa typing, multilocus sequence typing and staphylococcal cassette chromosome mec typing were applied to categorize the isolates. Phage typeability was 48.0% in MSSA and 91.3% in MRSA, highlighting the limitation of the PCR typing scheme when applied to PVL-positive MSSA. Five different PVL phages and two variants of a known phage were detected, the most prevalent being ΦSa2usa, recovered in 15 out of 48 (31.2%) isolates, and carried by both MSSA and MRSA belonging to CC8 and CC5. The recently described ΦTCH60 was recovered in four isolates. A PVL phage (ΦSa119) from an ST772 MRSA, that was not detected using the previous typing scheme, was sequenced, and new primers were designed for the identification of the icosahedral-head group II PVL phages present in ST772 and ST59 MRSA. A comprehensive PVL-phage typing can contribute to the understanding of the epidemiology and evolution of PVL-positive MSSA and MRSA.
Introduction
Since the end of the 1990s, community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) has been recognized as a major cause of skin and soft-tissue infections (SSTIs), and occasionally of serious invasive infections such as necrotizing pneumonia, in young subjects without apparent risk factors living in the community 1. In the USA a single CA-MRSA clone, designated USA300, is dominant, being responsible for the majority of the SSTIs seen in emergency departments 2. Different CA-MRSA clones have been detected in other areas of the world, such as ST80 in Europe and ST30 in Oceania 3; these clones, once apparently geographically restricted, have now been found in different continents, together with new emerging CA-MRSA lineages 4. With some exceptions, the majority of CA-MRSAs contain the genes for the Panton-Valentine leukocidin (PVL) 5, a bi-component, pore-forming toxin that causes lysis of leucocytes and subsequent tissue necrosis 6. PVL is also carried by some methicillin-susceptible S. aureus (MSSA) strains, sharing similar disease potential and epidemiological characteristics with CA-MRSA 7. In some countries, such as the UK, PVL-positive MSSAs are more common than MRSA in community-acquired infections 8.
PVL is encoded by two co-transcribed genes, lukS-PV and lukF-PV, carried by temperate phages lysogenized into S. aureus chromosome 9. PVL phages belong to Siphoviridae, a family of bacteriophages characterized by double-stranded DNA, with an icosahedral- or elongated-head morphology, and a non-contractile tail 10. To date, several PVL phages of the two morphological groups have been identified and sequenced 11-16.
In 2008, Ma et al. 15 developed a sequential PCR-based scheme for the detection and identification of the two morphological groups and of the five most common PVL phages, subsequently modified to include the PVL phage carried by USA 300 17.
Using this scheme, the distribution of PVL phages was investigated in various countries. In Japan, two types of PVL phages, Φ108PVL and ΦSa2958, were found to predominate 15, while in England and Wales the most common phage types were ΦPVL, ΦSa2usa and ΦSa2mw 17-19. In China, Φ108PVL was mostly recovered, although a specific phage type was only identified in nine out of 58 PVL-positive isolates 20.
Following the description in emerging CA-MRSA clones of new icosahedral-head PVL phages that escape typing by the previous scheme, a second morphological group of icosahedral-head PVL phages was identified 21, 22. Therefore, to date, PVL phages can be classified into three groups: the elongated-head group, icosahedral-head group I and icosahedral-head group II.
In Italy, a variety of genetically distinct CA-MRSA clones are present 23, and occurrence of PVL-positive MSSA in community-acquired infections has been reported 24, 25. The aim of this study was to type PVL phages carried by MSSA and MRSA isolated in Italy from community-acquired infections to better understand the epidemiology and the evolution of these pathogens.
Materials and Methods
Bacterial isolates
All the PVL-positive S. aureus isolates collected from April 2005 to May 2011 were examined in this study. Overall 48 PVL-positive S. aureus (25 MSSA, 52%, and 23 MRSA, 48%) were referred on a voluntary basis by 18 hospital laboratories located in seven different regions of north and central Italy. All the isolates had been obtained from severe infections or persistent or recurrent infections acquired in the community (Table 1).
| Isolate ID | Source | Year | ST | CC | MSSA/MRSA | SCCmec | spa type | Phage morphology | PVL-phage type |
|---|---|---|---|---|---|---|---|---|---|
| Sau017 | Necrotizing pneumonia | 2006 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau018 | Necrotizing pneumonia | 2007 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau016 | SSTI | 2006 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau065 | SSTI | 2009 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau116 | SSTI | 2010 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau130 | SSTI | 2010 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau150 | SSTI | 2010 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau161 | SSTI | 2011 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau164 | Nasal carrier | 2011 | 8 | 8 | MRSA | IV | t008 | Elongated-head | ΦSa2usa |
| Sau085 | SSTI | 2010 | 8 | 8 | MRSA | IV | t121 | Elongated-head | ΦSa2usa |
| Sau147 | SSTI | 2010 | 8 | 8 | MSSA | – | t4069 | Elongated-head | ΦSa2usa |
| Sau169 | Necrotizing fasciitis | 2011 | 8 | 8 | MSSA | – | t1265 | Elongated-head | ΦSa2usa |
| Sau151 | SSTI | 2010 | 1 | 5 | MRSA | V | t127 | Elongated-head | ΦSa2usa |
| Sau106 | Sepsis | 2009 | 1 | 5 | MSSA | – | t127 | Elongated-head | ΦSa2usa |
| Sau146 | SSTI | 2010 | 1 | 5 | MSSA | – | t127 | Elongated-head | ΦSa2usa |
| Sau019 | Necrotizing pneumonia | 2007 | 80 | 80 | MRSA | IV | t044 | Elongated-head | ΦSa2mw |
| Sau058 | SSTI | 2009 | 80 | 80 | MRSA | IV | t044 | Elongated-head | ΦSa2mw |
| Sau089 | Necrotizing fasciitis | 2010 | 80 | 80 | MRSA | IV | t044 | Elongated-head | ΦSa2mw |
| Sau149 | SSTI | 2010 | 80 | 80 | MRSA | IV | t044 | Elongated-head | ΦSa2mw |
| Sau574 | SSTI | 2009 | 80 | 80 | MRSA | IV | t044 | Elongated-head | ΦSa2mw |
| Sau021 | Necrotizing pneumonia | 2008 | 80 | 80 | MRSA | IV | t2453 | Elongated-head | ΦSa2mw |
| Sau086 | SSTI | 2010 | 22 | 22 | MRSA | IV | t005 | Icosahedral-head I | ΦPVL |
| SauL94 | SSTI | 2005 | 22 | 22 | MSSA | – | t005 | Icosahedral-head I | ΦPVL |
| SauL61 | Nasal carrier | 2005 | 954 | 22 | MSSA | – | t2336 | Icosahedral-head I | ΦPVL |
| SauL732 | Nasal carrier | 2005 | 956 | 30 | MSSA | – | t021 | Icosahedral-head I | ΦPVL |
| Sau015 | Necrotizing pneumonia | 2008 | 30 | 30 | MRSA | IV | t755 | Elongated-head | ΦTCH60 |
| Sau102 | SSTI | 2010 | 30 | 30 | MRSA | IV | t6955 | Elongated-head | ΦTCH60 |
| Sau022 | SSTI | 2008 | 5 | 5 | MRSA | IV | t319 | Elongated-head | ΦTCH60 |
| Sau056 | SSTI | 2009 | 88 | 88 | MSSA | – | t2310 | Elongated-head | ΦTCH60 |
| Sau004 | Osteomyelitis | 2007 | 121 | 121 | MSSA | – | t284 | Elongated-head | ΦSa2958 |
| Sau007 | SSTI | 2008 | 1210 | 121 | MSSA | – | t645 | Elongated-head | ΦSa2958 |
| Sau002 | Sepsis | 2006 | 30 | 30 | MSSA | – | t318 | Icosahedral-head I | Φ108PVL-V1a |
| Sau143 | SSTI | 2010 | 30 | 30 | MSSA | – | t021 | Icosahedral-head I | Φ108PVL-V2a |
| Sau009 | Nasal carrier | 2006 | 1210 | 121 | MSSA | – | t645 | Elongated-head | NT |
| Sau078 | SSTI | 2010 | 121 | 121 | MSSA | – | t308 | Elongated-head | NT |
| Sau080 | Necrotizing pneumonia | 2010 | 121 | 121 | MSSA | – | t159 | Elongated-head | NT |
| Sau114 | Osteomyelitis | 2010 | 51 | 121 | MSSA | – | t159 | Elongated-head | NT |
| Sau027 | SSTI | 2008 | 88 | 88 | MRSA | V | t2526 | Elongated-head | NT |
| Sau141 | SSTI | 2009 | 6 | 6 | MSSA | – | t7693 | Elongated-head | NT |
| Sau037 | Sepsis | 2009 | 152 | 152 | MSSA | – | t1172 | Elongated-head | NT |
| Sau136 | SSTI | 2010 | 30 | 30 | MSSA | – | t318 | Icosahedral-head I | NT |
| Sau139 | SSTI | 2010 | 30 | 30 | MSSA | – | t021 | Icosahedral-head I | NT |
| Sau142 | SSTI | 2010 | 1472 | 30 | MSSA | – | t665 | Icosahedral-head I | NT |
| Sau001 | ORL infection | 2006 | 1209 | 942 | MSSA | – | t1445 | Icosahedral-head I | NT |
| Sau008 | Nasal carrier | 2006 | 1209 | 942 | MSSA | – | t1445 | Icosahedral-head I | NT |
| Sau119 | SSTI | 2010 | 772 | 5 | MRSA | V | t7445 | Icosahedral-head II | ΦSa119 |
| Sau005 | SSTI | 2008 | 5 | 5 | MSSA | – | t1154 | Unknown | NT |
| Sau077 | Necrotizing pneumonia | 2010 | 1790 | 5 | MSSA | – | t311 | Unknown | NT |
- SSTI, skin and soft tissue infection; NT, non-typeable.
- a Variants of Φ108PVL (see text).
The isolates were assayed for the presence of PVL and of mecA by PCR according to previously published methods 23.
The control isolates for phage typing were: S. aureus 81/108 (Φ108PVL), JCSC2958 (ΦSa2958), A980470 (ΦSLT), MW2 (ΦSa2mw) and USA300 FPR3757 (ΦSa2usa). As no control was available for ΦPVL, strain SauL94 (this study) was used as a control.
Molecular typing
Staphylococcus aureus genomic DNA was extracted with the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). Molecular typing was performed by protein A (spa) typing and multilocus sequence typing (MLST); the staphylococcal chromosome cassette (SCC) mec was typed in MRSA isolates 23.
PVL phage detection
Phage typing was performed following the sequential scheme of Ma et al. 15 that allows classification of elongated-head group and icosahedral-head group I phages. Briefly, the three steps of the scheme detected the morphological phage group, the linkage with PVL genes and the specific phage type, respectively. The original scheme, allowing identification of five phage types, was extended to include two more phage types, ΦSa2usa 15, 17 and ΦTCH60 26 (Table 2).
| Primer use | Primer (pair) name | Target gene/locus | Reference | |
|---|---|---|---|---|
| I set | ICOS I morphology | portal-1F/portal-1R | por | 15 |
| tail-1F/tail-1R | mtp | 15 | ||
| ELONG morphology | portal-2F/portal-2R | por | 15 | |
| tail-2F/tail-2R | mtp | 15 | ||
| II set | ICOS I-PVL linkage | teil-ico-F/lukSR1 | mtp, lukS-PV | 15 |
| ELONG-PVL linkage | teilE-F2/lukSR1 | mtp, lukS-PV | 18 | |
| III set | ΦPVL and Φ108PVL | intF-2/PVL-aR/108-aR | int, JP030, ant | 15 |
| ΦSa2958 | intF-2/2958-aR | int, JP004 | 15 | |
| ΦSLT | intF-2/SLT-aR | int, ssb | 15 | |
| ΦSa2mw | intF-2/MW2-aR | int, cro | 15 | |
| ΦSa2usa | Sa2USA_F/USA2_R2a | phiSLT ORF484-like | 17 | |
| ΦTCH60 | intF-2/TCH60-aR | int, HMPREF0772_11656 | 26 |
- ICOS, icosahedral-head group I; ELONG, elongated-head group.
- a USA2_R2 (5′- CCTCAGCGCCATCACCAATA-3′), modified from primer Sa2USA_R 17.
If the third set of PCR assays did not identify any specific phage type, PVL phages were identified only on the basis of the morphological group. If the PCR assays of the scheme of Ma yielded negative results, the presence of an unknown PVL phage was confirmed using the primers designed by Boakes et al. 17, targeting the conserved left and right junctions of the PVL prophage with the bacterial chromosome.
Prophage induction in strain Sau119 and electron microscopy analysis
To detect the presence of a functional phage in strain Sau119, mitomycin induction was performed. Briefly, Sau119 was grown in 10 mL of BHI broth at 37°C to a turbidity corresponding to OD620 0.3. Mitomycin C (Sigma-Aldrich, St Louis, MO, USA) was added to final concentrations of 0.6 μg/mL and the culture was incubated at 37°C until lysis occurred, as shown by a decrease in turbidity. Cellular debris was removed by centrifugation at 16 000 g for 15 min at 4°C; the filtered supernatant was ultracentrifuged at 100 000 g for 2 h at 4°C. The pellet obtained was washed twice in PBS and resuspended in 200 μL of PBS; this represented the phage preparation and was stored at −80°C until used. For transmission electron microscopy the phage preparation was deposited on carbon-coated copper grids, negatively stained with 2% potassium phosphotungstate at pH 7.0 and examined in a Philips EM 208 electron microscope.
DNA sequencing of the Sau119 phage and design of a new typing scheme
Phage DNA was extracted from 100 μL of the phage preparation using the QIAamp DNAMini Kit (QIAGEN). The nucleotide sequence was determined by applying the 454-Genome Sequencer FLX procedure (http://454.com/applications/whole-genome-sequencing/) to libraries constructed on phage DNA, producing >600 bp fragment reads. The draft genome was assembled by the GS De Novo Assembler v.2.6 software (Roche Diagnostic, Monza, Milan, Italy).
Automated gene annotation was performed by the web-based Rapid Annotation using Subsystems Technology (RAST) service 27. Annotation of relevant genes was manually curated based on annotated staphylococcal phage genomes available in the GenBank database. The nucleotide sequence of the PVL phage carried by Sau119 (ΦSa119) has been deposited in GenBank under the Accession no. KJ596420.
Blast similarity search against the nucleotide collection database and whole-genome shotgun contigs database was performed at the NCBI website.
On the basis of the sequence of ΦSa119, an additional typing scheme consistent with the sequential scheme of Ma et al. 15, allowing identification of icosahedral-head group II PVL phages including ΦSa119, was designed (Table 3).
| Primer use | Primer name | Sequence (5′–3′) | Size of PCR product (bp) | Target gene | Reference | |
|---|---|---|---|---|---|---|
| I set | ICOS II morphology | PORT_3 | TTCGGAAGAATGATAGGTGC | 535 | por | This study |
| PORT_4 | TAATCCGCACCTCATTCCTTG | por | This study | |||
| TAIL_3 | TCAAAGAATGGCAGAAAGTGG | 842 | mtp | This study | ||
| TAIL_4 | ACTCTTGGTAACTAAAGCGAC | mtp | This study | |||
| II set | ICOS II-PVL linkage | TAIL_5 | AATTGGGATAGCAACGCAAGAGC | 10 728 | mtp | This study |
| LukS-R1 | ACGAAGTAGCAATAGGAGTGA | luks-PV | 15 | |||
| III set | ΦSa119 (ST772 phage) | intF-2 | ATGTTTTCGAGTTTTTGAGTTAG | 4918 | int | 15 |
| SA119Ant1 | AGTATTGAATTGTCGCTACCAGC | ant | This study | |||
| ST59 phage | intF-2 | ATGTTTTCGAGTTTTTGAGTTAG | 2965 | int | 15 | |
| repR | AGATGCTTACGTAAAGAAGG | rep | 21 |
Results
Molecular typing of the isolates
Molecular typing revealed the presence of 27 different spa types, 18 among MSSA, 11 among MRSA, and two shared by both MSSA and MRSA. Eighteen different STs were found, five of which were shared by both MSSA and MRSA. The most prevalent MSSA clonal groups were CC30 and CC121, each comprising six out of 25 isolates (24.0%). The most prevalent MRSA clonal group was CC8 (10 out of 23 isolates, 43.5%), followed by CC80 (six out of 23 isolates, 26.0%) (Table 1).
PVL-phage typing
Applying the expanded PCR scheme of Ma, it was possible to detect a PVL phage and determine at least the morphological phage group (elongated-head group or icosahedral-head group I) in 45 out of 48 isolates (93.7%). The large majority of the PVL phages (34 out of 48, 70.8%) were elongated-head types. A specific PVL-phage type was identified in 33 isolates (68.7%); phage typeability was 48.0% in MSSA (12 out of 25) and 91.3% in MRSA (21 out of 23) (Table 1).
The most common phage type was ΦSa2usa, recovered in 15 isolates (31.2%), both MRSA and MSSA belonging to CC8 and CC5 (Table 1). ΦTCH60 was recovered in four isolates (8.3%), three MRSA belonging to CC30 and CC5 and one MSSA belonging to CC88.
Among the icosahedral-head group I, two MSSA isolates belonging to CC30 (Sau002 and Sau143) yielded amplicons with primers designed to detect Φ108PVL (intF-2/108-aR, Table 2); however, the fragments obtained were larger than expected (c. 5.6 kb instead of 4.3 kb). Sequences of the 3′ end of the amplicons were different and had identity with regions of different PVL phages (Φ7247PVL (Genbank Accession no. AP01195) for Sau002 phage and tp310-1 (Genbank Accession no. NC_009761.2) for Sau143 phage). Therefore the two phages were considered different variants of Φ108PVL and designated Φ108PVL_V1 and Φ108PVL_V2, respectively (Table 1).
In 12 out of 48 isolates (25.0%), it was only possible to determine the morphological phage group and not the specific PVL-phage type.
With three isolates, including one MRSA belonging to ST772 (Sau119), all the PCR assays of the typing scheme yielded negative results, and accordingly the phage type was defined as unknown. In these isolates, the presence of a PVL phage that was integrated into the same chromosomal site of known PVL phages was confirmed by PCR.
Characterization of PVL phage ΦSa119
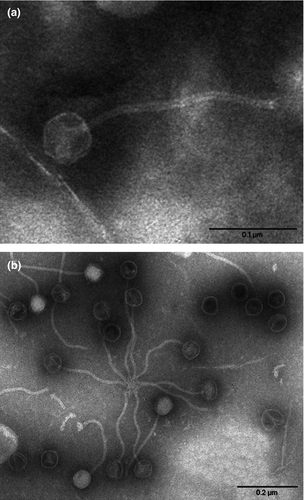
Electron microscopy of the supernatant of a mitomycin-induced culture of Sau119 revealed the presence of viral particles consisting of a small icosahedral head and a long flexible tail that are characteristic of bacteriophages belonging to the Siphoviridae family 28 (Fig. 1).
Following 454 sequencing of the purified PVL phage of Sau119, a unique contig accounting for the phage sequence with a 300-fold coverage, was obtained by GS-FLX gsAssembler software.
The phage, designated ΦSa119, was 42 600 bp in size with a GC content of 33.4%. A 29-bp core sequence of the attP site (5′- ACCATCACATTATGATGATATAATTAGTC -3′) was identified corresponding to the 29-bp sequence found at the L junction of the integrated phage (data not shown). The integration site of ΦSa119 was inside a region corresponding to the hypothetical gene with locus tag SACOL1532 in the S. aureus COL chromosome (Genbank accession no. CP000046).
Blast analysis of ΦSa119 sequence towards Whole Genome Shotgun database revealed 99% identity with a region inside genomes of ST772 isolates from India, Malaysia and Australia, corresponding to a PVL phage named ΦIND772PVL 22. As observed in ΦIND772PVL and other ST772 PVL phages 22, 29, ΦSa119 contains sea, the enterotoxin A gene, located in a region close to the PVL genes. Sequence analysis also revealed 94% nucleotide identity between ΦSa119 and PVL phages found in ST59 isolates, including Φ7247PVL (Genbank accession no. AP011956) 21, 30, 31.
PVL phages present in both ST772 and ST59 strains belong to the icosahedral-head group II 21, 22 and are not detected by the PCR scheme of Ma. Therefore a new scheme for PVL phages present in both ST772 and ST59 strains of the icosahedral-head group II was designed exploiting the identity between the structural genes of ST772 and those of ST59 PVL phages. For the first set of PCR assays the morphological group was detected by a duplex PCR targeting the structural genes por, encoding for the portal protein, and mtp, encoding a tail protein (Table 3). For the second set of PCR, the linkage between the tail gene and the PVL genes was detected with a primer pair targeting mtp and lukS. The confirmatory PCR for the ST772 phage type (third set of PCR) was developed using primers targeting int and ant, the putative antirepressor that appears to be specific for ΦSa119 and other ST772 phages (Table 3). ΦSa119 was identified as an ST772 phage by this new typing scheme, while the two MSSA isolates that carried unknown PVL phages according to the scheme of Ma, did not yield any positive result.
When one PVL-positive ST59 S. aureus control isolate (not included in this study) was examined by icosahedral-head group II typing, amplicons were obtained with primer sets 1 and 2, of the same size as those obtained with ΦSa119, indicating that the new scheme is able to detect the PVL phage present in ST59 at the morphological level, but no amplicon was obtained with the PCR specific for the ST722 phage type. A new PCR specific for the ST59 PVL-phage type was designed using published primers targeting int and rep (Table 3). This assay gave a positive result, allowing the identification of the ST59 PVL-phage type (not shown).
Discussion
This study reached a double result: a better characterization of PVL-positive MSSA and MRSA isolates from Italy, with the definition of the PVL phages harboured, and the expansion of the phage typing scheme of Ma et al. 15 to include new PVL-phage types.
In Italy, the most common PVL-positive MSSA clonal groups recovered were CC121 and CC30, in line with the study of Rasigade et al. 7 on a global collection of PVL-positive MSSA clinical isolates. Among PVL-positive MRSA, the most prevalent lineages were CC8 and CC80, as already reported in a previous study 23.
In this study, the original phage typing scheme of Ma has been expanded, allowing the identification of seven PVL phages, including ΦSa2usa 17 and the newly detected ΦTCH60 26. By the extended typing scheme, a specific PVL-phage type could be identified in 91% of MRSA and in 48% of MSSA. The limitation of the extended typing scheme for MSSA could be due to the higher genetic heterogeneity of PVL-positive MSSA in comparison with PVL-positive MRSA and to the fact that the scheme was originally developed for MRSA 15, 17, 18.
An additional typing scheme aimed at detecting PVL phages of a new morphological group (icosahedral-head group II) present in ST772 and ST59 clones that escapes detection by the scheme of Ma, has been proposed. This typing scheme, which should be applied in the case of negativity of the PCR scheme of Ma, expands the detection of icosahedral-head phage types, allowing identification of the PVL-phage types present in ST772 and ST59, clones emerging in Asia and other parts of the world 21, 22, 30. In Italy these clones are still rare; the isolate carrying ΦSa119 was probably imported into our country, because it was isolated from a man of Indian nationality.
This study highlights the variety of the PVL phages recovered in a single European country: ΦSa2usa, initially identified in the USA300 clone, was the most common, but five other distinct PVL-phage types, including two variants of Φ108PVL and ΦSa119 of ST772, were identified.
A lineage-specificity of PVL phages was noted in our study, irrespective of the methicillin-resistant status of the isolates, in line with observations previously reported 15, 17-19. The occurrence of a single phage in a clone comprising both MSSA and MRSA suggests that the acquisition of PVL phage is likely to have occurred before the acquisition of SCCmec that generated MRSA 32-34. The recovery of ΦSa2958, previously associated with MRSA belonging to CC30, in two MSSA belonging to CC121 is the first report of PVL phages present in this most prevalent lineage of PVL-positive MSSA 7. ΦTCH60, recently identified in CC30 26, was also detected in CC5 and CC88 isolates.
The large variety of phages detected suggests that in Europe and in other areas where PVL-positive MSSA and MRSA belong to several different clones 26, the characterization of PVL phages could be informative for the epidemiology of S. aureus and contribute to outbreak investigation.
We have an example in our collection: MSSA strain SauL94 (ST22), carrying ΦPVL, was the epidemic strain responsible for a prolonged community and hospital outbreak of SSTI in northern Italy 24. During the outbreak, many staff members carried the PVL-positive epidemic strain but one of the carriers was colonized by a different strain, belonging to ST30 (SauL732), that also contained ΦPVL. Because ΦPVL is uncommon in ST30 isolates, we hypothesized that SauL732 acquired ΦPVL from the epidemic strain.
One limitation of this study is that the identification of PVL phages is based on PCR assays that target only a portion of the phage genome. Because phages are prone to recombination events, positivity by PCR should not be taken as indication of a specific phage but rather of a type or family of PVL phages with variable genomic portions. This occurrence has been shown in our study when amplicons from two isolates obtained with primers for Φ108PVL were sequenced and showed identity with fragments of other PVL phages. Next-generation sequencing, which is becoming increasingly available and affordable, will generate more robust data on the diversity of PVL phages, allowing a more comprehensive epidemiological picture of PVL-positive S. aureus.
Acknowledgements
We thank the Italian clinicians and microbiologists who provided the clinical isolates, and Teruyo Ito, Angela Kearns, Frederic Laurent and Michèle Bes for providing the positive control strains. This publication made use of the Multi Locus Sequence Typing website (http://www.mlst.net) at Imperial College London developed by David Aanensen and funded by the Wellcome Trust.
Transparency Declaration
This work was supported in part by the CCM project ‘Sorveglianza dell'antibiotico-resistenza in comunità, nelle infezioni trasmesse dagli alimenti ed in quelle di origine zoonosica’ of the Italian Ministry of Health and the Italian FLAGSHIP ‘InterOmics’ project (PB.P05) CNR- MIUR. None of the authors has declared any conflict of interests.




