Characterization of the first report of Mycobacterium timonense infecting an HIV patient in an Ecuadorian hospital
Abstract
Mycobacterium timonense is a non-tuberculous mycobacteria (NTM) described in southern France in 2009, and to our knowledge, not reported again as a human pathogen in indexed literature. The aim of this work was to characterize the first clinical isolate of M. timonense in Ecuador. Time of growth, biochemical tests, thin layer growth test, PCR-RFLP analysis of the hsp65 gene and MALDI-TOF spectra analysis were not able to identify the species. The species identification was achieved through sequencing of rrs, hsp65 and rpoB genes. The results highlight the necessity to set up a sequencing method to identify emerging NTM in Ecuadorian clinical facilities.
The Mycobacterium genus comprises around 157 species, some of which are highly pathogenic to both humans and animals 1. Non-tuberculous mycobacteria (NTM) are diverse and ubiquitous in the environment 2. Human-human transmission is rare, while most of the infections are acquired by aerolization, ingestion or contact with injured tissue 3. The mycobacterium avium complex (MAC) is the principal NTM associated with human infections in immunocompromised and elderly patients; this complex includes Mycobacterium avium, Mycobacterium intracellulare and recently Mycobacterium chimaerera, Mycobacterium colombiense, Mycobacterium arosiense, Mycobacterium vulneris, Mycobacterium marseillense, Mycobacterium bouchedurhonense and Mycobacterium timonense 4. The methods commonly used for NTM species identification in Ecuadorian hospitals, such as biochemical tests, thin layer growth test, PCR-RFLP analysis (PRA) of the Hsp65 gene and MALDI-TOF, are not able to identify the recently described species of the MAC, such as M. timonense, for which sequencing is necessary. To our knowledge, since the description of M. timonesnse in southern France in 2009, no cases of human infection associated with this pathogen have been reported in the indexed literature. We describe the first isolation and characterization of M. timonense from an HIV patient.
Colonies were isolated from a synovial fluid sample and membrane biopsy from a 44-year-old man with AIDS, classification C3 (viral load, 207 000 copies/mL and CD4: 3) with highly active antiretroviral therapy (HAART). The patient had arthritis of the right elbow so he was treated at the Traumatology Service of Vozandes Hospital in Quito, Ecuador. The samples were inoculated in Lowenstein-Jensen slants and the MGIT 960 system, because the Ziehl-Neelsen stain (AFB) was positive in both samples. After 10 days of incubation, the MGIT detected growth, while small colonies were observed with the Lowenstein-Jensen method. A routine thin layer growth and biochemical tests were carried out manually.
The DNA of the isolate (denominated NTMZ&Z1) was obtained using the High Pure PCR template preparation kit (Roche Diagnostic, Schweiz, Switzerland). DNA was subjected to PRA analysis 5. Separate aliquots of PCR amplification of the 441 bp of the hsp65 gene were digested with BstEII and HaeIII, and resolved in 2% agarose gel. Restriction patterns observed were compared with Chimara et al. 5.
MALDI-TOF analysis was carried out in a Microflex LT MALDI-TOF spectrometer (Bruker Daltonics Inc., Bremen, Germany). The spectrum was examined in a MALDI-TOF Biotyper v. 3.1.
The PCR fragments of 1500 bp of the rrs, 441 bp of the hsp65 and 733pb of the rpoB gene were sequenced at the Macrogen (Korea) facilities. Sequences were run against MAC sequences deposited in the GenBank. Sequence alignment and a UPGMA tree (bootstrap with 1000 replicates) of the concatenated sequences (2542 bp: 1428 bp of the rrs, 707 bp of the rpoB and 407 bp of the hsp65 gene) were built using Geneiuos v.7.0.6.
The isolate was identified as M. chelonae/M. peregrinum by routine biochemical tests. Thin layer test showed a unique growth pattern (Fig. 1a) and MALDI-TOF analysis showed a high score (2.095), but was not able to differentiate between M. chimera and M. intracellulare (Fig. 1b). The PRA pattern was very similar to M. chimaera and M. intracellulare according to the algorithm described by Chimara et al. 5 and M. marseillense according to Wallace et al. 6 (Fig. 1c). Finally, the sequences of rrs (KJ364652), rpoB (KJ364653) and hsp65 (KJ364654) genes showed 100% identity with M. timonense in Blast analysis and a UPGMA tree generated using concatenated sequences (Fig. 1d, Table 1).
| Method | Identification | Comment |
|---|---|---|
| Biochemical tests | M. chelonae/M. peregrinum | NTMZ&Z1 biochemical profile does not match exactly with any profile 11a |
| Thin layer | ND | New growth pattern |
| PRA | M. chimera/M. intracellulare | Mycobacterium timonense PRA pattern is not described in Chimara et al.'s 2008 PRA algorithm |
| MALDI-TOF | M. chimera/M. intracellulare | MALDI-TOF Biotyper version 3.1 does not include M. timonense mass spectra |
| Assembly of rrs, rpoB and hsp65 gene sequences (2542pb) | M. timonense | Identify M. timonense with bootstrap 100% and identity 100% |
- a Biochemical tests: growth speed, growth in NaCl 5%, growth at 45°C, pigmentation, tween 80 hydrolysis, utilization of citrate, alkaline phosphatase, pirazinamidase and urease.
- PRA, PCR-RFLP analysis; MALDI-TOF, matrix-assisted laser desorption/ionization-time-of-flight; ND, non determined.
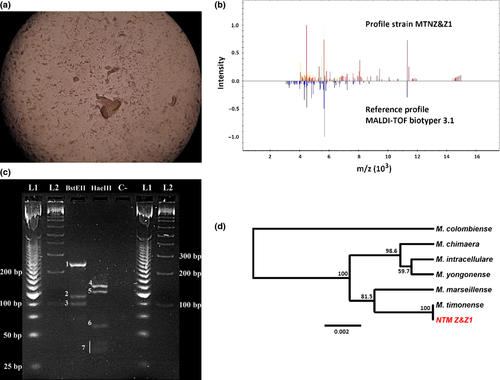
The mycobacterium avium complex is primarily pulmonary pathogens that affect individuals who are immune compromised (AIDS, immunosuppressive chemotherapy and hairy cell leukaemia). In this clinical setting, the MAC has been associated with osteomyelitis, tenosynovitis, synovitis and disseminated disease involving the lymph nodes, the central nervous system, the liver, the spleen and the bone marrow. The MAC comprises the most common pathogens causing infection by NTM in patients with AIDS. M. avium is isolated in more than 95% of patients with AIDS who develop MAC infections 7, 8. Still, the epidemiology of distinct species of the MAC is not completely understood 9. Recently, nine species have been described within the MAC, all of them with clinical implications. However, lack of genotyping techniques available has been one of the main reasons why there are very few data available on species from this complex 9. This could be illustrated by the results obtained using the most common methods applied in Ecuadorian clinical facilities, which could not distinguish between several species of Mycobacterium (Table 1, Fig. 1).
Single and concatenated sequences of rrs, rpoB and hsp65 genes strongly suggest that the isolate found in the present study was M. timonense. Bootstrap values had high support in all nodes in the UPGMA tree, confirming that these genes provide high resolution for identification of species of the MAC. A similar strategy has been used in order to identify emerging pathogen Mycobacterium species 10 (Fig. 1). The lack of success in Mycobacterium identification by healthcare facilities could be obscuring our knowledge of the real distribution and epidemiology of NTM strains at a global scale. This is relevant as understanding the distribution and epidemiology would be the first step in trying to assess differences in virulence between some of the newly described members of the MAC.
This is one of the first global reports since the species was described in 2009 11. We described an opportunistic infection caused by M. timonense in an AIDS patient, who was treated with ethambutol, clarithromycin and levofloxacin for 6 months. The outcome was favourable. This report has relevance from an epidemiological perspective because it suggests that MAC species in HIV patients are a potential source of infections and should be routinely screened for and identified by sequencing. Countries in the region have found novel MAC species (Mycobacterium colombiense) causing infections in immunocompromised individuals 12. As far as we know, most hospitals do not routinely screen for and identify NTM in our country, and the very little information that is generated in terms of epidemiology of this group of bacteria has not been published in the indexed literature. Moreover, non-sequence-based methods, which are commonly used in most Ecuadorian hospitals, are not able to distinguish between related NTM species. In this context, our findings suggest that many standard methods for identifying Mycobacteria could be misleading for accurate species identification of the MAC and encourage us to recommend the use of sequence methods to identify these pathogens in local clinical facilities.
Acknowledgements
All molecular assays were supported by Zurita&Zurita Laboratorios. Biochemical tests and thin layer growth test were carried out in Hospital Vozandes Quito, Ecuador. The MALDI-TOF spectrum was obtained in the Centro de Investigaciones Microbiológicas. The authors are grateful to Yolanda Izurieta, Cecibel González, Andrea Carrera and Pedro Barba, for the technical assistance in laboratory procedures. The primary results of this study were presented at the VIII Congreso Ecuatoriano de Patología Clínica/Medicina de Laboratorio,16–18 October 2013.
Transparency Declaration
The authors declare no conflicts of interest.