Establishment of a system to quantify wild-type herpes simplex virus–induced cell–cell fusion reveals a role of N-glycosylation of HSV-1 envelope glycoprotein B in cell–cell fusion
Ayano Fukui and Yuhei Maruzuru contributed equally to this study.
Abstract
Wild-type herpes simplex virus (HSV) strains infrequently mediate cell–cell fusion in cell cultures and barely induce large multinucleated cells. In this study, we established a system to quantify infrequent cell–cell fusion induced by wild-type HSV strains. The established system clarified that the HSV-1 envelope glycoprotein B and its N-glycosylation at asparagine at position 141 were required for efficient cell–cell fusion. This study provides a link between cell–cell fusion induced by wild-type HSV-1 and viral pathogenesis in vivo.
Abbreviations
-
- g
-
- glycoprotein
-
- HSV-1
-
- herpes simplex virus 1
-
- HSV-2
-
- herpes simplex virus 2
-
- Vero-TagRFP
-
- Vero cells stably expressing TagRFP
-
- Vero-Venus
-
- Vero cells stably expressing Venus
INTRODUCTION
Herpes simplex virus 1 and 2 (HSV-1 and -2) are human pathogens that cause various diseases, including encephalitis, keratitis, and mucocutaneous and skin diseases such as herpes labialis, genital herpes, and herpetic whitlow.1 Similar to other enveloped viruses, membrane fusion is a critical step in the replication of HSV. Thus, infection of host cells with HSV is initiated by fusion of the viral envelope with the host cell membrane, followed by delivery of the nucleocapsid into the host cell.2 Envelope glycoprotein B (gB), gD, and a complex of gH and gL are thought to be sufficient for fusion upon HSV entry, based on cell-based fusion assays showing that co-transfection of genes encoding gB, gD, gH, and gL into cells expressing an HSV entry receptor(s) results in massive fusion of the cells with one another.3 Infection with envelope viruses such as measles virus, human respiratory syncytial virus, severe acute respiratory syndrome coronavirus 1 and 2, and human immunodeficiency virus causes extensive cell–cell fusion in cell cultures, resulting in large multinucleated cells.4 Similarly, HSV infection induces cell–cell fusion, whereas cell–cell fusion caused by wild-type HSV strains is infrequent in cell cultures, and large multinucleated cells are barely observed.5-7 However, wild-type HSV strains induce low but consistent cell–cell fusion,6, 7 and large multinucleated cells are frequently observed in herpetic lesions in humans.4 Although these observations suggest that cell–cell fusion contributes to HSV pathogenesis, the relevance of HSV-induced cell–cell fusion in viral replication and pathogenesis in vivo remains largely unknown. Furthermore, infrequent cell–cell fusion induced by wild-type HSV strains has barely been focused and studied. In this study, we established a system to quantify cell–cell fusion induced by wild-type HSV strains. Using this system, we demonstrated that the N-glycan of HSV-1 gB, which was previously reported to be significant for viral neurovirulence and replication in the central nervous system in mice,8 was required for cell–cell fusion induced by wild-type HSV-1 in cell culture.
MATERIALS AND METHODS
Cells and viruses
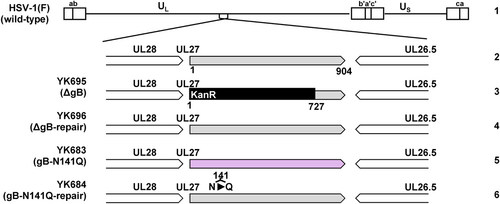
Vero, and Plat-GP cells have been described previously.9, 10 Wild-type HSV-1(F), wild-type HSV-2(186), YK695(ΔgB), YK696(ΔgB-repair), YK683(gB-N141Q), and YK684(gB-N141Q-repair) (Figure 1) have also been previously described.8, 11, 12
Plasmids
To construct pMX-Venus and pMX-TagRFP, Venus or TagRFP open-reading frame sequences were amplified using PCR from pBS-Venus13 or pTagRFP-N114 and inserted into the BamHI/NotI site of pMX,15 respectively.
Establishment of Vero cells stably expressing Venus or TagRFP
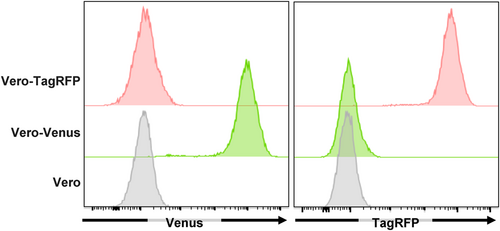
To generate Vero-Venus (Vero cells stably expressing Venus) or Vero-TagRFP (Vero cells stably expressing TagRFP) cells, Vero cells were transduced with supernatants of Plat-GP cells co-transfected with pMDG16 and pMX-Venus or pMX-TagRFP, and selected by sorting of Venus+ or TagRFP+ cells using an FACS Melody cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA).
Cell fusion assays
Vero-Venus and Vero-TagRFP cells were cocultured at a 1:1 ratio in 12 well plates 1 day before infection. Thereafter, cells were infected with the selected viruses at an MOI of 1. After adsorption for 1 h, the inoculum was removed and the cell monolayers were overlaid with medium 199 containing 10% FCS. At 24 h postinfection, infected cells were detached using trypsin, washed once with PBS supplemented with 2% FCS, and analyzed with a CytoFLEX S (Beckman Coulter, Brea, CA, USA). Data were analyzed using FlowJo (version 10.8.1) software (Becton Dickinson). Flow cytometry data depicted single cells, defined by sequential removal of doublets according to forward scatter-area × forward scatter-height and side scatter-area × side scatter-height.
Statistical analysis
One-way analysis of variance followed by Tukey's test was used for multiple comparisons. A P value < 0.05 was considered statistically significant. GraphPad Prism 8 (GraphPad Software, Boston, MA, USA) was used for the statistical analysis.
RESULTS
Establishment of a quantitative system for wild-type HSV-induced cell–cell fusion
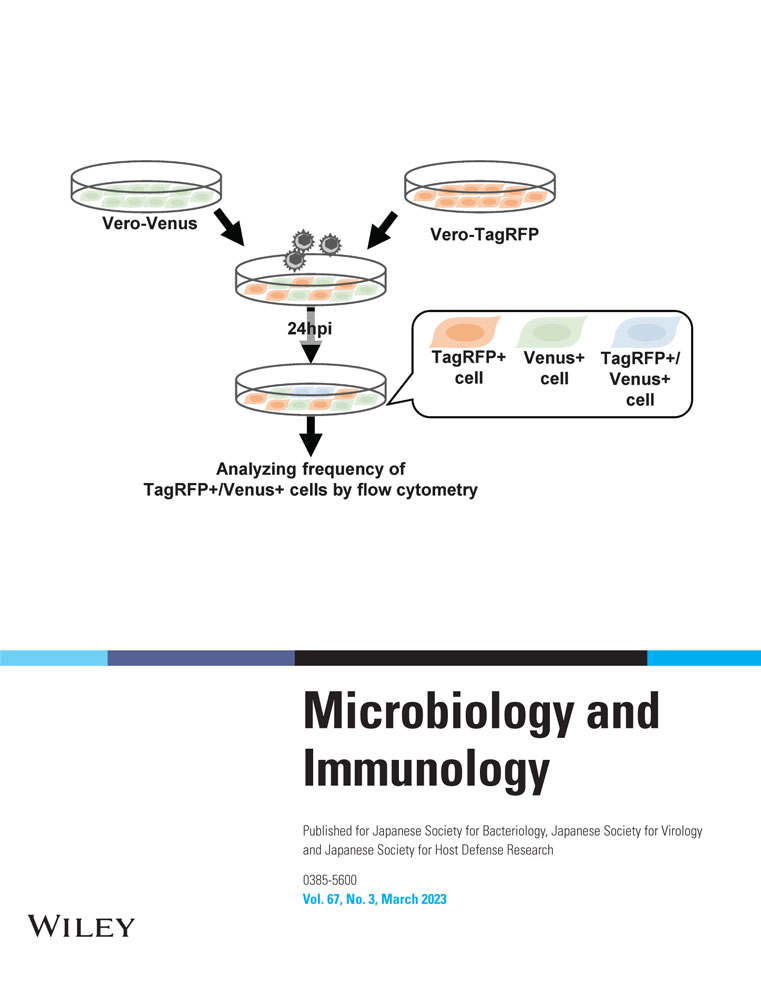
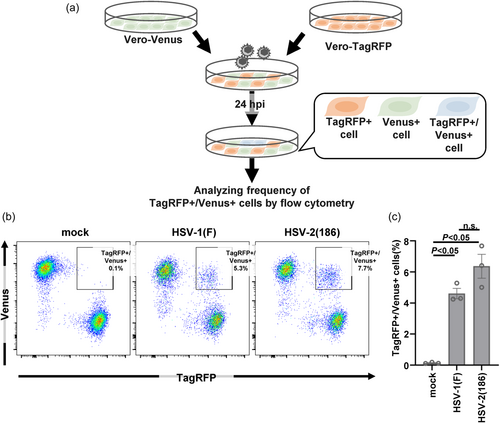
To quantify the infrequent cell–cell fusion caused by wild-type HSV strains in cell culture, we generated Vero cells that stably expressed the monomeric fluorescent protein VenusA206K17 or TagRFP18 and designated them as Vero-Venus or Vero-TagRFP cells, respectively (Figure 2). The Vero-Venus and Vero-TagRFP cells were mixed in a 1:1 ratio. One day after cocultivation, the cells were mock infected or infected with wild-type HSV-1(F) or wild-type HSV-2(186) at an MOI of 1 for an additional 24 h and then subjected to flow cytometry. As the cells positive for both Venus and TagRFP fluorescence resulted from cell–cell fusion between Vero-Venus and Vero-TagRFP cells, virus-induced cell–cell fusion was evaluated by analyzing the frequency of TagRFP+/Venus+ cells using flow cytometer (Figure 3a). As shown in Figure 3b,c, although TagRFP+/Venus+ cells were barely detectable (0.11%) in mock-infected cells, the frequency of TagRFP+/Venus+ cells significantly increased to 5.3% and 7.7% in cells infected with wild-type HSV-1(F) and wild-type HSV-2(186), respectively. These results indicate that this established system could detect infrequent cell–cell fusion caused by wild-type HSV-1 and HSV-2.
Effect of gB on HSV-1–induced cell–cell fusion
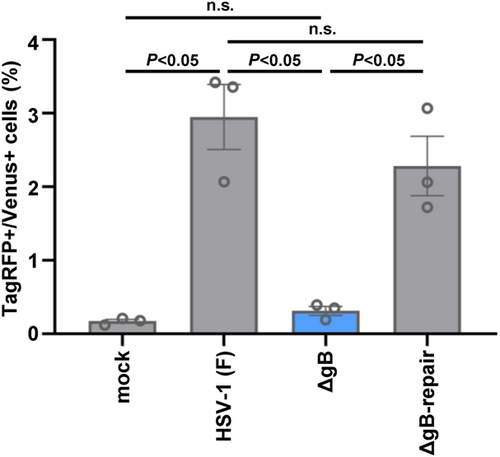
It has been reported that cell–cell fusion induced by other envelope viruses involves virus-encoded fusion proteins.4 HSV-1 gB is a class III fusion glycoprotein that plays an essential role in the fusion of the envelope with host cell membrane during virus entry.19 However, whether gB is required for infrequent cell–cell fusion induced by wild-type HSV strains remains unclear. To address this issue, cocultured Vero-Venus and Vero-TagRFP cells were mock-infected or infected with wild-type HSV-1(F), a gB-null mutant ΔgB,8 or its repaired virus ΔgB-repair8 and subjected to flow cytometry. As shown in Figure 4, the frequency of TagRFP+/Venus+ cells in ΔgB infection was significantly lower than that in wild-type HSV-1(F) or ΔgB-repair infection. These results indicate that gB is required for efficient cell–cell fusion caused by wild-type HSV-1. Notably, the frequencies of cells in ΔgB infection were as low as those in mock-infected cells, suggesting that gB is critical for cell–cell fusion caused by wild-type HSV-1.
Effect of the N-glycan at gB Asn-141 on cell–cell fusion induced by HSV-1
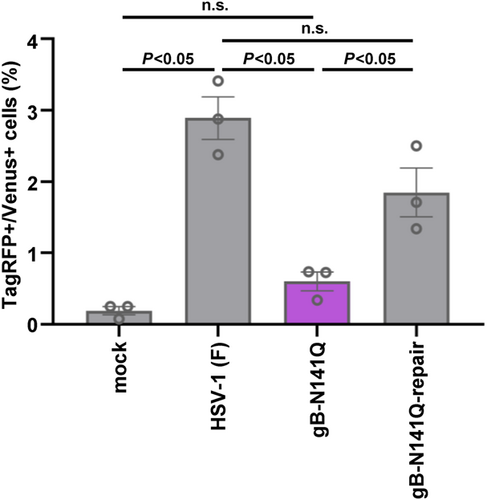
We previously reported that the N-glycan at gB Asn-141 is required for efficient cell surface expression of gB, without affecting its subcellular localization.8 To examine whether N-glycans at gB Asn-141 play a role in the cell–cell fusion induced by wild-type HSV-1, cocultured Vero-Venus and Vero-TagRFP cells were mock infected or infected with wild-type HSV-1(F), a recombinant virus gB-N141Q in which gB Asn-141 was substituted with glutamine,8 or the repaired virus gB-N141Q-repair, followed by flow cytometry. As shown in Figure 5, the frequency of TagRFP+/Venus+ cells in gB-N141Q infection was significantly lower than that in wild-type HSV-1(F) or gB-N141Q-repair infection as observed with ΔgB infection. These results suggest that N-glycosylation at gB Asn-141 is required for efficient cell–cell fusion caused by wild-type HSV-1.
DISCUSSION
Virus-mediated cell–cell fusion is conserved among certain envelope viruses belonging to different families, suggesting its importance in various viral infections.4 HSV-induced cell–cell fusion has long been investigated using syncytial variants of laboratory stocks, which aberrantly cause extensive cell–cell fusion in cell cultures, resulting in large multinucleated cells.4, 5 However, syncytial mutants rarely occur in clinical samples of HSV and emerge during the propagation of cultured cells, suggesting that these mutations artificially occur in cell cultures and reduce viral replication and pathogenesis in vivo. Considering these findings, the significance of cell–cell fusion in HSV infection should be investigated using wild-type strains. However, cell–cell fusion induced by wild-type HSV strains remains largely unstudied, probably owing to the low frequencies of cell–cell fusion.
In this study, we established a system to quantify infrequent cell–cell fusion induced by wild-type HSV strains. This system appeared to be easy to operate and sensitive to detect cell–cell fusion. The reporter cells (Vero-Venus and Vero-TagRFP cells) generated for this system were derived from Vero cells, which are susceptible to a variety of viruses. Therefore, the established system with reporter cells could be applicable for quantitative analyses of cell–cell fusion induced by other viruses.
Using the established system, we demonstrated that gB and its N-glycosylation at Asn-141 are important for cell–cell fusion induced by wild-type HSV-1 in cell culture. Notably, we previously reported that glycosylation at gB Asn-141 is required for the proper cell surface expression of gB in HSV-1–infected Vero cells.8 Furthermore, N-glycosylation was significant for HSV-1 neurovirulence and replication in the central nervous system of mice, although it played no obvious role in viral replication in Vero and neuroblastoma SK-N-SH cells.8 Collectively, these observations suggest that N-glycosylation at HSV-1 gB Asn-141 acts in cell–cell fusion induced by wild-type HSV-1 by regulating proper cell surface expression of gB and that this regulation might be involved in HSV-1 pathogenesis in vivo. This study sheds light on the role of cell–cell fusion induced by wild-type HSV-1 and provides a link between HSV-1–mediated cell–cell fusion and viral pathogenesis in vivo. Further studies to clarify the significance and mechanisms of cell–cell fusion induced by wild-type HSV strains are necessary and interesting and are currently under way in this laboratory.
ACKNOWLEDGMENTS
Foundation.We thank Risa Abe, Keiko Sato, Tohru Ikegami, and Yui Muto for providing technical assistance. This study was supported by Grants for Scientific Research and Grant-in-Aid for Scientific Research (S) (20H05692) from the Japan Society for the Promotion of Science (JSPS); grants for Scientific Research on Innovative Areas (21H00338, 21H00417, 22H04803); a grant for Transformative Research Areas (22H05584) from the Ministry of Education, Culture, Science, Sports and Technology of Japan; a PRESTO grant (JPMJPR22R5) from Japan Science and Technology Agency (JST); grants (JP20wm0125002, JP20wm0225009, JP20wm0225017, JP22fk0108640, JP22gm1610008, JP223fa627001) from the Japan Agency for Medical Research and Development (AMED); grants from the International Joint Research Project of the Institute of Medical Science, the University of Tokyo; and grants from the Takeda Science Foundation, the Uehara Memorial Foundation, and the Mitsubishi
DISCLOSURE
The authors have no conflict of interest to disclose regarding this work.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.