Genetic characterization of an avian H4N6 influenza virus isolated from the Izumi plain, Japan
ABSTRACT
An influenza A virus of H4N6 subtype was isolated from the Izumi plain, Japan, in 2013. Genetic analyses revealed that two viral genes (M and NS gene segments) of this isolate were genetically distinct from those of the H4N6 virus isolated from the same place in 2012. Furthermore, three viral genes (PB2, PB1 and M gene segments) of this isolate share high similarity with those of the North American isolates of 2014. These results suggest a high frequency of genetic reassortment of avian influenza viruses in Asian waterfowl and intercontinental movements of avian influenza viruses via migratory waterfowl.
Abbreviations
-
- HA
-
- hemagglutinin protein
-
- M
-
- matrix protein
-
- NA
-
- neuraminidase
-
- NP
-
- nucleoprotein
-
- NS
-
- nonstructural protein
-
- PA
-
- polymerase acid protein
-
- PB1
-
- polymerase basic protein 1
-
- PB2
-
- polymerase basic protein 2
Influenza A viruses, a member of the family Orthomyxoviridae, have eight-segmented genomes. The segmented viral genome allows influenza A viruses to exchange individual gene segments with their counterparts by co-infection of a single cell with multiple virus strains, a process called genetic reassortment 1, 2, leading to rapid evolution and emergence of genetic variants 3, 4.
Migratory waterfowl, especially wild ducks, are considered to be natural reservoirs for influenza A viruses 5, 6. because avian influenza viruses are mainly transmitted through the fecal–oral route 5, 7, the World Organization for Animal Health recommends using fecal samples from migratory waterfowl for surveillance of avian influenza 8. In addition, avian influenza viruses are reportedly carried along the flyways of migratory waterfowl between their nesting and overwintering sites 9-12. Japan is a known part of the East Asia/Australian flyway, which has an area that overlaps with the Pacific and Mississippi America flyways 6. Therefore, several avian influenza viruses reassorted with multiple lineages have been isolated in Japan 13-15.
The Izumi plain in Japan is known to be an overwintering site for over 10,000 endangered cranes and more than 20,000 wild ducks. Although these migratory waterfowl share the same overwintering site, their nesting sites and flyways are known to be different 16. Therefore, avian influenza viruses may infect certain groups of wild ducks, be brought to the Izumi plain and be transmitted to other groups of birds. Indeed, we have isolated various subtypes of avian influenza viruses, including highly pathogenic avian influenza viruses, from both migratory waterfowl and the environmental water of the Izumi plain 16-18. To further clarify the ecology of avian influenza viruses, we attempted to isolate influenza A viruses from wild duck fecal samples collected from the Izumi plain during the 2013/2014 winter season.
To isolate influenza A viruses, we collected 1000 samples of wild duck feces from the Izumi plain in 200 tubes (five fecal samples per tube) on 9 November 2013. Because numerous influenza A viruses were isolated from the environmental water of the Izumi plain in November 17, we collected the wild duck fecal samples from the same cranes’ roosting area during November and suspended pooled fecal samples in transport medium, as described previously 17. After filtration with 0.22 μm pore filters, we inoculated the supernatants from each fecal sample into the allantoic cavities of ten-day-old embryonated chicken eggs. After incubation for 48 hrs, we collected the allantoic fluid samples and performed hemagglutination assays using 0.5% chicken red blood cell suspensions (Nippon Bio-Test Laboratories, Saitama, Japan). We then found that one of the allantoic fluid samples showed hemagglutination activity (8 HA unit) and detected influenza A viral M gene from the sample by RT RT-PCR-based influenza viral gene detection. These findings indicate that we had isolated an influenza A virus from the wild duck feces collected at the Izumi plain.
To characterize the isolated influenza A virus, we determined the nucleotide sequences of the viral genes. The sequence data revealed that our isolate was classified under the H4N6 subtype (Table 1). It is worth noting that our isolate, A/duck/Kagoshima/KU-6/2013 (H4N6), possesses a monomeric basic residue at the HA cleavage site, indicating low pathogenicity 19. We phylogenetically analyzed the determined sequences of the viral genes with their representative counterparts. We aligned the nucleotide sequences with the MUSCLE program 20 and constructed the phylogenetic trees for each viral gene by using the neighbor-joining method based on maximum composite likelihood in MEGA 7 software 21 with a bootstrapping set of 1000 replicates. All the genes of our isolate belonged to the same clusters as did the genes of viruses isolated from migratory waterfowl in Asian countries in each phylogenetic tree (Figs. 1 and 2), indicating that A/duck/Kagoshima/KU-6/2013 (H4N6) is one of the low pathogenic avian influenza viruses circulating among Asian waterfowl.
| Isolated virus | Gene | Accession No.† | Closest relative‡ | Identity (%) |
|---|---|---|---|---|
| A/duck/Kagoshima/KU-6/2013(H4N6) | PB2 | EPI969350 | A/northern pintail/Alaska/830/2014 (H3N6) | 99.47 |
| PB1 | EPI969351 | A/northern pintail/Alaska/870/2014 (H3N8) | 99.34 | |
| PA | EPI969352 | A/duck/Vietnam/LBM48/2011 (H3N2) | 99.44 | |
| HA | EPI969353 | A/duck/Hunan/S11090/2012 (H4N6) | 98.64 | |
| NP | EPI969354 | A/duck/Vietnam/LBM48/2011 (H3N2) | 98.86 | |
| NA | EPI969355 | A/duck/Fujian/S2169/2012 (H4N6) | 99.22 | |
| M | EPI969356 | A/northern pintail/Alaska/870/2014 (H3N8) | 99.69 | |
| NS | EPI969357 | A/duck/Japan/11OG1032/2011 (H5N2) | 99.16 |
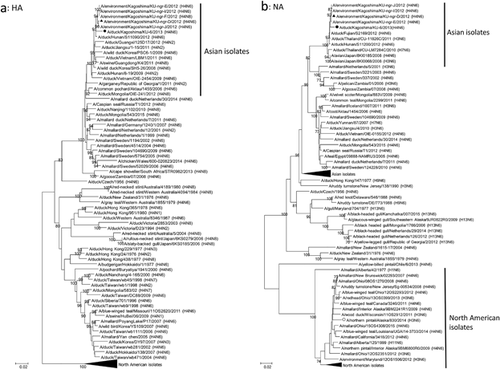
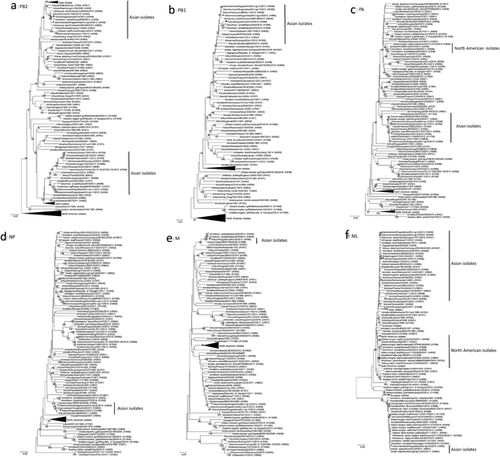
In November 2012, during the season before the season described in this study, we isolated an H4N6 avian influenza virus, A/environment/Kagoshima/KU-ngr-D/2012 (H4N6), from the environmental water of the Izumi plain 18. The phylogenetic trees revealed that the M and NS genes from A/duck/Kagoshima/KU-6/2013 (H4N6) are genetically distinct from those of A/environment/Kagoshima/KU-ngr-D/2012 (H4N6) (Fig. 2e,f), whereas the other genes of both isolates belonged to the same clusters (Figs. 1 and 2). These findings indicate that A/duck/Kagoshima/KU-6/2013 (H4N6) is not a direct progeny of A/environment/Kagoshima/KU-ngr-D/2012 (H4N6), but a reassortant with the other avian influenza virus(es) circulating among Asian waterfowl. These findings indicate genetic reassortment of avian influenza viruses in Asian waterfowl. Because we treated pools of five fecal samples as single specimens, it is possible that we inoculated multiple viruses from different fecal samples into single embryonated chicken eggs. However, the calculated probability that one of the 200 tested specimens contained multiple viruses is quite low (< 0.5%), suggesting that our isolate was unlikely to be have been generated through genetic reassortment in the inoculated chicken eggs.
Intriguingly, the PB2 gene of A/duck/Kagoshima/KU-6/2013 (H4N6) shares high similarity (99.47%) with the counterpart of a North American isolate, A/northern pintail/Alaska/830/2014 (H3N6) 21 (Fig. 2a and Table 1). The PB1 and M genes from A/duck/Kagoshima/KU-6/2013 (H4N6) also share high (>99.34%) similarity with their counterparts of another North American isolate, A/northern pintail/Alaska/870/2014 (H3N8) 21 (Figs. 2b,e and Table 1). These findings suggest that the PB2, PB1 and M gene segments of a close relative(s) of A/duck/Kagoshima/KU-6/2013 (H4N6) were carried from Asia to North America in 2014. In addition, the HA gene from A/northern pintail/Alaska/830/2014 (H3N6) and both HA and NA genes from A/northern pintail/Alaska/870/2014 (H3N8) also belong to the same phylogenetic clusters as each counterpart from the Asian waterfowl isolates (data not shown). Eventually, we determined that four (the PB2, PB1, HA and M) and five (the PB2, PB1, HA, NA and M) gene segments from A/northern pintail/Alaska/830/2014 (H3N6) and A/northern pintail/Alaska/870/2014 (H3N8), respectively, are of Asian lineage (Figs. 1 and 2). These findings suggest that these two isolates are reassortants between avian influenza viruses circulating among Asian and North American waterfowl and generated in 2014. Although low pathogenic avian influenza viruses in Asia and North America are thought to be evolving independently, previous studies have also reported intercontinental movements of Asian viruses and/or their gene segments to North America in 2014 22, 23 and earlier 24-26.
In the 2014/2015 winter season, we isolated eight highly pathogenic H5N8 avian influenza viruses that belong genetically to three groups; the isolates in two of the groups are genetically similar to the European and North American H5N8 viruses isolated during the same season 16. Therefore, the ancestor(s) of these H5N8 viruses are believed to have been introduced to Siberia, which is one of the major nesting sites of migratory waterfowl, at the end of the 2013/2014 winter season, where they infected not only Asian, but also European and North American migratory waterfowl populations, and thus spread to each of these regions in the subsequent season 27, 28. Our isolation of A/duck/Kagoshima/KU-6/2013 (H4N6), the PB2, PB1 and M genes of which are the closest relatives of the counterparts from two North American isolates, A/northern pintail/Alaska/830/2014 (H3N6) and A/northern pintail/Alaska/870/2014 (H3N8), at the Izumi plain in the 2013/2014 winter season in Japan, also support this phenomenon of intercontinental movements of avian influenza viruses in 2014.
In conclusion, we have isolated a low pathogenic H4N6 avian influenza virus from the Izumi plain in Japan. The M and NS genes are genetically distinct from those of an H4N6 virus that we isolated from the same place one season earlier. In addition, the PB2, PB1 and M genes share high similarity with those of the North American viruses isolated one season later. These findings highlight genetic reassortment and intercontinental movements of avian influenza viruses in migratory waterfowl.
ACKNOWLEDGMENTS
This work was supported by grants from the Project of the NARO Bio-oriented Technology Research Advancement Institution (Integration Research for Agriculture and Interdisciplinary Fields and R&D matching funds on the field for Knowledge Integration and Innovation) and by a grant for the contracted research activity of crane conservation from the City of Izumi, Japan. This research was commissioned by the Kagoshima Crane Conservation Committee.
DISCLOSURE
The authors declare no conflict of interest and confirm that this manuscript is not currently under consideration by any other journal.