A novel 111-nucleotide duplication in the G gene of human metapneumovirus
ABSTRACT
In 2017, novel human metapneumovirus (HMPV) A2b subgroup strains with a 111-nucleotide duplication in the G gene was detected by the present team. These strains were related to previously identified HMPV A2b strains with a 180-nucleotide duplication; however, they appeared to be different strains, produced by an independent duplication event. The recent evolution of HMPV suggests that careful monitoring of this virus is required.
Abbreviations
-
- 111nt-dup
-
- 111-nucleotide duplication
-
- 180nt-dup
-
- 180-nucleotide duplication
-
- 37aa-dup
-
- 37-amino-acid duplication
-
- A2b111nt-dup HMPV
-
- HMPV A2b strains with a 111nt-dup in the G gene
-
- A2b180nt-dup HMPV
-
- HMPV A2b strains with a 180nt-dup in the G gene
-
- Fusion
-
- G, glycol-
-
- HMPV
-
- human metapneumo virus
-
- RSV
-
- respiratory syncytial virus
Human metapneumovirus, a member of the family Pneumoviridae, is a major causative agent of acute respiratory infections, especially in children and older adults. This virus was discovered in 2001 1; however, serological studies have shown that it had been circulating, worldwide, for more than six decades 1. The viral genome is non-segmented negative-sense RNA and encodes nine proteins, including three surface glycoproteins: F, SH (small hydrophobic) and G proteins. On the basis of its nucleotide sequence and antigenic variations, HMPV is divided into two groups, A and B 2; each viral group being further divided into two subgroups: A1 and A2 in Group A and B1 and B2 in Group B 3, 4. Further detailed sequence analyses of HMPV strains have suggested two clades, A2a and A2b, in the A2 subgroup 5. Recently, unique A2b180nt-dup HMPV were detected in Japan and Spain 6, 7. Our work has suggested that HMPV-A2b180nt-dup emerged between 2011 and 2013 6. In the present study, we further report other novel A2b111nt-dup HMPV.
The analyses presented here were performed with the approval of the ethics committee of Yokohama City Institute of Public Health. In accordance with the National Epidemiological Surveillance of Infectious Diseases, instituted by Infectious Diseases Control Law in Japan, 1505 clinical specimens (throat swabs and nasal secretions) were collected from patients with upper or lower acute respiratory infections in Yokohama city, Kanto area, Japan, between January 2013 and June 2017 (data from 1306 of these samples from January 2013 to June 2016 have been reported previously) 6. The RNA of these clinical samples was extracted using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's protocol and subjected to multiplex RT-PCR using a Seeplex RV15 OneStep ACE Detection kit (Seegene, Seoul, South Korea) to detect 15 major respiratory viruses. The 122 samples that tested positive for HMPV were then further analyzed. The sequence of the HMPV G gene was determined as described previously 6. The cDNA of the HMPV F-gene was amplified with a PrimeScript II High Fidelity One Step RT–PCR Kit (TaKaRa Bio, Otsu, Japan) and subjected to a direct sequencing using a 3500 Genetic Analyzer (Applied Biosystems, Waltham, MA, USA). The nucleotide sequence data reported in the present study have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession numbers LC270124, LC275891, LC275892 and LC316180.
In 2017, three novel HMPV A2b strains with 111nt-dup in the G gene (A2b111nt-dup HMPV) were identified by the present team. Before 2016, no A2b111nt-dup HMPV strain had been detected 6, 7 (Table 1). Relevant basic and clinical characteristics of the three individuals infected with A2b111nt-dup HMPV are summarized in Table 2. All three presented with high fever and respiratory symptoms, as is typical of HMPV infection. The A2b111nt-dup HMPV strains in Patients 2 and 3 (HMPV/Yokohama.JPN/P8943/2017 and HMPV/Yokohama.JPN/P8945/2017, respectively) shared the same nucleotide sequence in the F and G genes. Although no direct epidemiological link was identified between Patient 2 (2.2 years old) and Patient 3 (1.9 years old), older sisters of these patients who attended the same kindergarten had also developed upper respiratory infections.
| Year of virus detection | |||||||
|---|---|---|---|---|---|---|---|
| Subgroup | Duplication | 2013† | 2014† | 2015† | 2016† | 2017 | Total |
| A2a | None | 4 | 1 | 1 | 0 | 0 | 6 |
| A2b | None | 11 | 3 | 12 | 3 | 0 | 29 |
| 111nt-dup‡ | 0 | 0 | 0 | 0 | 3 | 3 | |
| 180nt-dup§ | 0 | 3 | 10 | 2 | 0 | 15 | |
| B1 | None | 2 | 2 | 1 | 8 | 8 | 21 |
| B2 | None | 14 | 6 | 0 | 15 | 0 | 35 |
| Total | 31 | 15 | 24 | 28 | 11 | 109 | |
- †Data from January 2013 to June 2016 were reported in our previous paper (6); ‡111-nucleotide duplication; §180-nucleotide duplication.
| Patient | 1 | 2 | 3 |
|---|---|---|---|
| Age (years) | 3.8 | 2.2 | 1.9 |
| Sex | Male | Female | Male |
| Date of onset† | 30/4/2017 | 24/5/2017 | 28/5/2017 |
| Date of sample collection† | 2/5/2017 | 27/5/2017 | 28/5/2017 |
| Clinical diagnosis | Pharyngitis | Bronchitis | Bronchitis |
| Body temperature (°C)‡ | 39.9 | 39.5 | 39.6 |
| Hospitalization | No | No | No |
- †dd/mm/yyyy; ‡maximum body temperature.
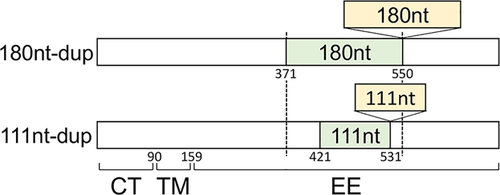
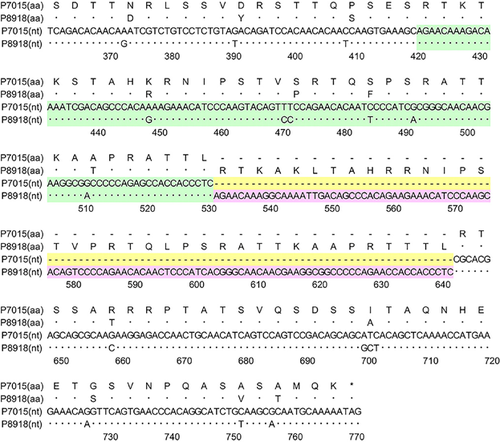
Figure 1 shows the structures of the G genes of A2b180nt-dup and A2b111nt-dup HMPV strains schematically. Figure 2 shows the nucleotide and deduced amino acid sequences at the duplicated region of the G gene of a representative A2b111nt-dup HMPV strain and a classical HMPV strain. 111nt-dup is a duplication of the 111 nucleotides at nucleotide positions 421–531, whereas 180nt-dup is a duplication of the 180 nucleotides at nucleotide positions 371–550 (the first nucleotide of the initiation codon was defined as nucleotide position 1). Compared with the classical A2b strain, which lacks these duplications, the A2b111nt-dup HMPV strain has a 37aa-dup in the C-terminal half-region of the extracellular ectodomain. Glycosylation site predictions performed with the NetNGlyc 1.0 8 and NetOGlyc 3.1 programs 9 indicated that A2b111nt-dup HMPV strains have acquired 11–12 additional potential acceptor sites for O-linked sugars. No potential acceptor site for N-linked sugars has been added by the 37aa-dup.
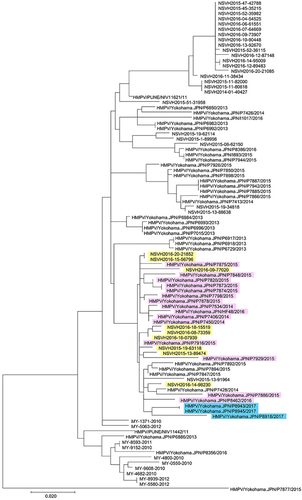
Phylogenetic analyses were performed with MAFFT (v7.304b) 10 and MEGA (v7.0.20) 11 software, as described previously 6. In the phylogenetic tree of the G gene, the three A2b111nt-dup HMPV strains formed a cluster together with 15 A2b180nt-dup HMPV strains and four classical A2b HMPV strains (HMPV/Yokohama.JPN/P7892/2015, HMPV/Yokohama.JPN/P7894/2015, HMPV/Yokohama.JPN/P7847/2015 and HMPV/Yokohama.JPN/P7428/2014) detected in Yokohama city, Japan, between 2014 and 2016 (Fig. 3), suggesting that these 180- and 111-nt-dup have occurred in recent years. A similar cluster was also found in the phylogenetic tree of the F gene (Fig. 4).
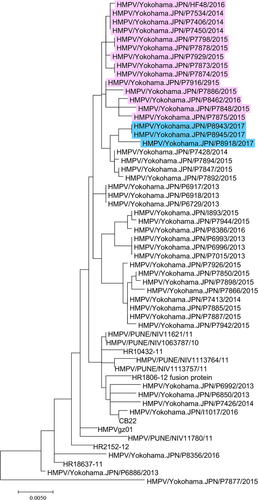
In summary, after 2014, at least two types of duplicated sequence insertion (180nt-dup and 111nt-dup) were detected in the G gene of major endemic HMPV strains; 111nt-dup being detected in 2017. Although their sizes differ, the locations of the duplicated regions are similar for 180nt-dup and 111nt-dup (nucleotide regions 371–550 and 421–531, respectively; Fig. 1). Although A2b180nt-dup and A2b111nt-dup strains may have a common ancestral classical A2b strain, the genome structures of the two strains suggest that they independently acquired their nt-dup in this region of the G gene. The reason for the recent evolution in the G gene of HMPV is unknown; however, similar nt-dup have also been reported in a related virus, human orthopneumovirus (human RSV), a member of the family Pneumoviridae 12, 13. RSV strains with nt-dup have rapidly spread globally and are currently the predominant strains in many countries 14, 15. These data suggest that careful global monitoring of both HMPV and RSV are necessary.
ACKNOWLEDGMENTS
We thank the staff of the clinics and hospitals that collected the specimens and clinical information. We are also grateful to all members of the Yokohama City Institute of Public Health for their technical support and dedicated assistance. This work was partly supported by the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development, AMED and the Japan Foundation for Pediatric Research (No 15-004).
DISCLOSURE
The authors have no conflicts of interest to declare.