U2AF1S34/Q157 Variants Detected in cis Arise Sequentially in an MDS Patient With Serial Sequencing Spanning 18 Years
Approximately 50% of patients with myelodysplastic syndromes (MDS) have mutations in splicing factor genes, including SF3B1, SRSF2, and U2AF1.1, 2 Variants found in the U2AF1 gene are highly specific and usually heterozygous, affecting either the S34 or Q157 residues, which are located in the three cysteine residues and one histidine residue (C3H1 or CCCH) zinc fingers of the protein.3 Rarely, these U2AF1 variants can be found to co-occur in individuals with myeloid malignancies.
Recently, in a study exploring allele-specific differences in splicing factor mutations, Taylor et al4 identified 9 patients with dual U2AF1S34/Q157 variants. Focusing on a single patient in their cohort, they demonstrated co-occurrence of the variants in cis with preservation of the wild-type allele in single-sequenced cells, and hypothesized these occurred around the same time. We present an additional case of a patient in our previously published cohort5 found to have U2AF1S34/Q157 variants also in cis, but sequentially acquired, as demonstrated in serial sample sequencing results spanning 18 years.
A nonsmoking 74-year-old male with history of scoliosis, hernia repair, and gastroesophageal reflux disease, presented in September 2018 with normocytic anemia and thrombocytopenia, and a hypercellular bone marrow (BM) with erythroid hyperplasia and dysplasia, 25% ring sideroblasts (RS), megakaryocytic dysplasia, 3% myeloid blasts and a normal karyotype. Findings were consistent with MDS with RS and multilineage dysplasia. Targeted next-generation sequencing (NGS) was performed as previously described,5 and variant analysis using the Oncomine Myeloid Assay (“Oncomine” filter chain; Thermo Fisher Scientific, Waltham, MA) revealed the following variants and allele fractions: U2AF1S34F (47%); U2AF1Q157R (33%); and SETBP1D868N (48%). Supplementary analysis with a custom pipeline (as previously described5) revealed additional variants: ETV6G375R (43%) and PRPF8D1598N (55%).
Notably, the patient presented with macrocytic anemia 16 years earlier (2002), with BM demonstrating similar erythroid dysplasia, with lack of RS and no significant megakaryocytic dysplasia. At that time, the patient was identified as having probable MDS (46,XY) based on their dysplastic features but was lost to Hematology follow-up until 2018. The archived BM sample was retrieved and sequenced at the time of the September 2018 assessment, revealing that the U2AF1S34F variant was present at the time of initial BM in 2002. This suggests a possible long evolution of MDS, with additional somatic variants leading to newly observed dysplastic features. Furthermore, this patient had 2 additional BM assessments conducted to monitor potential disease evolution, in May (MDS with excess blasts 1 [MDS-EB-1]) and August (MDS-EB-2) of 2019, with NGS revealing the presence of additional variants at each time point (Table 1). Treatment initially involved supportive care with darbepoetin, red cell, and platelet transfusions. The patient began azacitidine treatment in September 2019, completing 16 cycles before expiring in December 2020. We also sequenced archived BM slides from February 2020, revealing that variant allele frequencies (VAFs) of some previously detected variants decreased following azacitidine treatment, while others increased (Table 1).
| Variant | 2002-01 | 2018-09 | 2019-05 | 2019-08 | 2020-02 |
|---|---|---|---|---|---|
| U2AF1S34F | 42.6% | 47.0% | 46.0% | 44.3% | 47.4% |
| ETV6 G375R | 0.25%a | 42.6%b | 28.5%b | 1.71%a | 0.38%a |
| PRPF8 D1598N | — | 55.4%c | 25.9%c | — | 0.58%a |
| SETBP1 D868N | — | 48.0% | 53.0% | 65.2% | 89.3% |
| U2AF1Q157R | — | 33.0% | 45.0% | 48.3% | 49.1% |
| ETV6 R105G | — | 0.39%a | 14%b | 21.3%b | 43%b |
| KRAS G13D | — | 0.28%a | 2.7%b | 14.0% | 42.5% |
| KRAS G12S | — | — | 4.2% | 22.5% | 4.3% |
| ASXL1 R693* | — | 1.79%a | 2.8% | 23.4% | 3.6% |
| PTPN11 E76G | — | 0.98%a | — | 2.6%b | 0.25%a |
- aDetected by IGV manual inspection only.
- bDetected by Thermo Fisher's newly released “Oncomine Extended” filter,6 but not detected by “Oncomine” filter alone.
- cNot detected by “Oncomine Extended” filter, nor “Oncomine” filter.
- — = not detected; BM = bone marrow; IGV = Integrative Genomics Viewer; VAFs = variant allele frequencies.
The inferred clonal evolution for this 18-year span is modeled in Figure 1A, including VAFs (Figure 1B) based on manual curation of all variants in Integrative Genomics Viewer (version 2.11; Broad Institute, Cambridge, MA),7 blood cell counts, and BM blast percentages (Figure 1C–G) taken at each time point. This analysis revealed that several variants were present at time points of BM assessment but were not reported because they fell below the validated limit of detection for the assay (Table 1). Reporting of these variants at that early time point may have modified prognosis and patient care. This highlights the importance of defining clinically relevant limits of detection, especially in the context of monitoring myeloid clonal dynamics and disease evolution.
Additionally, with the U2AF1S34/Q157 variants in 2018 and 2019 samples, we sought to determine whether they occurred in cis. Three archived genomic DNA (gDNA) samples from this period were subjected to long-range polymerase chain reaction (PCR) using the LA PCR Kit, v2.1 (Takara Bio, Kusatsu, Japan; RR013B), with primers designed to include both U2AF1 variants of interest (NM_006758.2: Chr21:43104346; p.Ser34Phe and Chr21: 43094667; p.Gln157Arg), with the following sequences: forward 5’-AAACAGAAACGACACCTATGCCAA-3’; reverse 5’-AAAAGTCTGTGTCATGTTTCTGTGAGG -3’. PCR products were run on a 0.4% agarose gel with the GeneRuler High Range DNA Ladder (Thermo Fisher Scientific; SM1351). DNA was extracted using QIAEX® II Gel Extraction Kit (Qiagen, Venlo, The Netherlands; 20021). Amplified, extracted DNA was sequenced on a MinION device (Oxford Nanopore Technologies [ONT]; Oxford, United Kingdom) using the standard manufacturer recommended protocol. Briefly, amplicons from each sample were barcoded using the native barcoding kit (ONT, SQK-NBD112.96) prior to library preparation with the ligation sequencing kit (ONT, SQK-LSK109). MinION sequencing was carried out for 48 hours with live basecalling enabled. The resultant FAST-Quality (FASTQ) files were aligned using Minimap28 and reads containing specific variants were extracted using the tool Binary Alignment Map Query Language (BAMQL).9
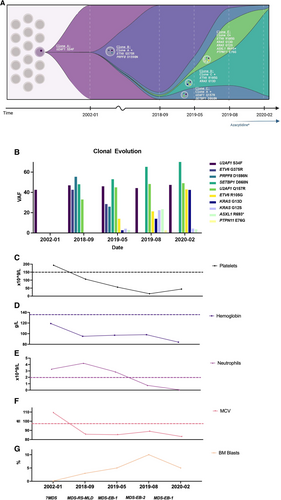
Clonal evolution and clinical counts for patient 1 over time. (A), Clonal evolution and inferred clonal dynamics at each NGS time point. *Azacitidine treatment started 2019-09, administered ~every 4 wk, on days 1, 2, 3, 4, 5, 8, 9; last treatment given in 2020-12 prior to expiry. (B), VAFs for variants at each NGS time point. (C–G), Clinical counts at each time point. Dashed lines represent lower normal limit for platelets (C), hemoglobin (D), and neutrophils (E); and the upper limit for MCV (F). (G), BM. BM = bone marrow; MCV = mean corpuscular volume; MDS-EB-1 = myelodysplastic syndrome with excess blasts 1; MDS-EB-2 = myelodysplastic syndrome with excess blasts 2; MDS-RS-MLD = myelodysplastic syndrome with ring sideroblasts and multilineage dysplasia; NGS = next-generation sequencing; VAFs = variant allele frequencies.
Nanopore sequencing on all 3 samples (2018-09, 2019-05, and 2019-08) revealed that the U2AF1S34/Q157 variants occurred in cis, with preservation of the wild-type allele. While Taylor et al4 proposed that these variants occurred around the same time, our novel findings reveal that the U2AF1 variants in this patient were acquired sequentially. While we cannot exclude co-occurrence of U2AF1S34/Q157 variants, we provide evidence that they can occur sequentially.
Furthermore, given that the U2AF1S34 variant was present in 2002 at a similar VAF (near 50%) to that seen in later samples, we sought to confirm that it was not present in the germline. We retrieved an archived, formalin-fixed, paraffin-embedded colonic biopsy sample. gDNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, 56404) and sequenced as previously described.5 The U2AF1S34 variant was only detected at a VAF of 7%, consistent with leukocyte infiltration commonly seen in systemic tissues.
In our cohort, 2/168 (1.2%) patients had U2AF1S34/Q157 variants. While patient 1 has been described herein, a second patient had an overlapping MDS/myeloproliferative neoplasm, with U2AF1S34/Q157 and signal transduction-related mutations: NRASG12D and PTPN11T507L. Unfortunately, there was no available DNA of sufficient quality to confirm whether their U2AF1S34/Q157 variants occurred in cis. However, this patient expired 1 year after diagnosis, indicating aggressive myeloid disease. Given that patient 1 also acquired RAS pathway mutations later in their disease evolution (KRASG13D and KRASG12S), surveillance for these emerging clones may be warranted in patients with existing U2AF1 mutations.
Importantly, the PRPF8D1598N variant that went undetected by Oncomine filters involved an RNA splicing factor gene not included in the analysis by Taylor et al.4 However, based on our inferred clonal evolution (Figure 1A), it may co-occur with the U2AF1S34F variant, potentially tolerated in early stages but outcompeted by clones with a more prominent advantage later in disease progression (ie, additional U2AF1Q157R variant). Future studies should explore the potential functional impact of the specific PRPF8D1598N variant on RNA splicing, such as through RNAseq of serial samples, given that its functional and clinical significance remain unclear.10 If the variant displays evidence of pathogenicity, the potential co-occurrence of PRPF8 variants with other splicing factor mutations and the potential impact on clonal dynamics and myeloid disease evolution should also be further explored.
The extended follow-up period and serial sequencing for patient 1 provides important insight into clonal dynamics, allowing progression to be closely monitored over time. However, many of these variants were present but were not reported based on the limitations of standard NGS testing. Error-corrected and single-cell sequencing represent important technological advances that may improve sensitivity for detection of emerging clones; future prospective studies should thus explore the potential use of these new technologies for more precise detection of clonal dynamics and evolution. Insights provided by these highly sensitive techniques may ultimately facilitate more informed decisions regarding patient care, including when to start more intensive treatments.
ACKNOWLEDGMENTS
The authors acknowledge the contributions to sequencing of the KHSC Molecular Genetics lab, especially Laura Semenuk and Dr Henry Wong; the input provided by Department of Pathology and Molecular Medicine Bioinformatician, Dr Kathrin Tyryshkin; and Dr Reina Ditta at McMaster University for DNA extraction of FFPE sample.
AUTHOR CONTRIBUTIONS
CKF and MJR planned and designed the study. CKF, AJMM, FB, PZ, and DL collected data. CKF and MJR wrote the article. All others commented on and approved the article.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
The authors acknowledge the Southeastern Ontario Academic Medical Organization (SEAMO); the Ontario Institute for Cancer Research (OICR); and the Canadian Institutes of Health Research (CIHR) for funding first-author PhD candidate, CKF with a Banting and Best Canada Graduate Scholarship (Doctoral).