Rapid Generation of Leukemogenic Chromosomal Translocations in Vivo Using CRISPR/Cas9
The authors declare no conflicts of interest.
Supplemental Digital Content is available for this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.hemaspherejournal.com).
Abstract
Supplemental Digital Content is available in the text
A wide range of chromosomal rearrangements have been identified in human cancers especially in leukemia.1, 2 Murine models that recapitulate chromosomal translocations are essential for understanding the determinants of malignancies and as a basis for preclinical development of rational therapeutic methods. Genetically engineered murine models involving chromosomal translocation genes have been generated using transgenic approaches or gene knockin.3, 4 However, the closest model is the creation of bona fide chromosomal translocations, which recapitulate the physiologic levels of the fusion genes, the reduced dosage of the wild-type alleles and the contribution of the reciprocal products of the translocations. This has been achieved in vivo using the Cre-loxP system in embryonal stem cells,5 which is labor intensive and time consuming. The CRISPR/Cas9 system is a versatile tool for genomic engineering. Transduction of target cells with vectors encoding Cas9 and sgRNAs targeting two genomic loci causes two DNA double strand breaks (DSBs), which could induce chromosomal rearrangements as a result from non-homologous end joining-based DNA repair.6 This strategy has been used to model oncogenic chromosomal rearrangements in murine7-10 or human11 cells in vivo. However, given the challenges of delivering CRISPR/Cas9 vectors into hematopoietic cells, the penetrance of leukemogenesis has been shown with low frequency in vivo.11 Here we establish an efficient methodology to generate chromosomal translocations in mice hematopoietic cells based on a vector design enabling convenient lentiviral delivery of multiple sgRNAs to hematopoietic cells from a Cas9 expressing mouse strain.
The KMT2A (MLL) gene constitutes a hotspot for chromosomal translocations in human leukemia, resulting in over 135 different leukemia-associated KMT2A gene fusions.2 KMT2A-MLLT1 defines a leukemia subtype with poor outcome. This translocation frequently constitutes the sole genomic lesion to drive the leukemia,12 providing a good model to establish this approach. We designed a strategy to induce Kmt2a-Mllt1 translocations in murine hematopoietic cells using the CRISPR/Cas9 system followed by transplantation into lethally irradiated recipient mice (Fig. 1A-B). Given the large size of Cas9, it is challenging to transduce murine hematopoietic cells with lentiviral vectors coding Cas9 together with sgRNAs. We therefore took advantage of the Cas9 knockin mouse,13 in which the Cas9 and a GFP reporter are expressed constitutively in the whole body. Using a single lentiviral vector to deliver two U6 promoter-sgRNA cassettes is an efficient way to induce two DSBs simultaneously.14 We constructed a lentiviral vector coding sgRNA and a fluorescent marker (sg_shuttle_RFP657, Supplementary Fig. 1A, http://links.lww.com/HS/A96) with special designs: (1) The small size of the backbone with minimum interval sequences between each modules improves the virus titer; (2) The isocaudomer sites flanking the sgRNA cassettes facilitates the combination of multiple single sgRNAs from individual vector into one vector (Supplementary Fig. 1B, http://links.lww.com/HS/A96). We firstly cloned several individual sgRNAs targeting the equivalent introns of Kmt2a and Mllt1 that are orthologue regions in the human leukemia translocation (Fig. 1A-B). The editing efficiency of each sgRNA was firstly validated in NIH 3T3 cells stably expressing Cas9 (Supplementary Fig. 1C, http://links.lww.com/HS/A96), then the most efficient sgRNAs targeting Kmt2a and Mllt1 loci were combined into one vector (sg_ K-M). Transduction of this vector coding two sgRNAs in NIH 3T3 cells generated the Kmt2a-Mllt1 translocation efficiently (Supplementary Fig. 1D, http://links.lww.com/HS/A96).
We then applied this approach to target primary murine hematopoietic cells (Fig. 1C). Since the original Cas9 knockin mouse has a mixed background of B6/129/FVB,13 we crossbred the Cas9 mice with the inbred C57BL/6 mice and used the F1 progenies (Cas9GFP+/−) as the bone marrow donors. Given the C57BL/6 haploidentical setting, these cells can reconstitute the hematopoietic system in lethally irradiated C57BL/6 recipient mice without inducing graft-versus-host disease. Transduction of these bone marrow cells with the sgRNA vector (sg_ K-M) lead to a detectable genomic rearrangement between the Kmt2a and Mllt1 locus at 4 days after transduction in ex vivo cultures (Fig. 1D). The expression of both Kmt2a-Mllt1 and the reciprocal Mllt1-Kmt2a fusions were detected by RT-PCR and verified by sequencing (Fig. 1D-E). Droplet Digital PCR (ddPCR) performed on genomic DNA from unsorted cells detected a copy number ratio of 0.003 of the Kmt2a-Mllt1 with respect to the wild type Kmt2a, implicating that ∼0.6% of the cells harbor the translocations (Fig. 1F). Thus this strategy is fast and practical to generate de novo translocations in primary murine hematopoietic cells.
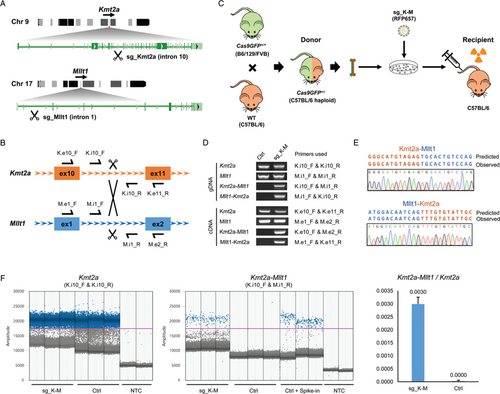
Induction of the Kmt2a-Mllt1 rearrangement in murine hematopoietic cells using the CRISPR/Cas9 system. (A) Schematic of the t(9;17) involving the Kmt2a and Mllt1 loci. Introns targeted by sgRNAs are indicated. (B) Schematic of the Kmt2a and Mllt1 loci with the locations of the primers used for genotyping. (C) Strategy for the generation of the Kmt2a-Mllt1 rearrangement in murine bone marrow cells and transplantation experiments. (D) PCR genotyping of cultured bone marrow cells from Cas9GFP+/− mice transduced with or without sgRNA vectors (sg_K-M). PCRs were performed on genomic DNA (gDNA) or cDNA from bone marrow cells ex vivo cultured for 4 days after transduction. (E) Sequence of the PCR products from cDNA showing the Kmt2a-Mllt1 and Mllt1-Kmt2a junctions. (F) ddPCR quantification of Kmt2a and Kmt2a-Mllt1 frequencies in gDNA from bone marrow cells ex vivo cultured for 4 days after transduction of sgRNA vectors. Signal amplitudes of amplicon-positive (blue) and -negative (gray) droplets for the detection of Kmt2a and Kmt2a-Mllt1 (left and middle) and the copy number ratio of Kmt2a-Mllt1 with respect to Kmt2a in Ctrl and sgRNA transduced groups (right, data are presented as mean with SD). Primer pairs for detections are indicated in the parenthesis; NTC, no template control; Ctrl+Spike-in, control gDNA spike-in with synthetic artificial DNA templates of Kmt2a-Mllt1.
Transplantation of the engineered cells into lethally irradiated C57BL/6 mice lead to engraftment of a GFP+ population, which can be detected in peripheral blood at 4 weeks after transplantation. Three out of nine mice displayed a marked increasing of the RFP657+ populations in the myeloid population over time, which suggested the sgRNA-transduced population to become dominant in vivo (Fig. 2A). By 120 days after transplantation, 100% of the Kmt2a-Mllt1 mice presented signs of sickness and were sacrificed (Fig. 2B). Post-mortem examination of Kmt2a-Mllt1 mice confirmed the development of leukemia with predominant myeloblasts in the peripheral blood, splenomegaly and extensive infiltration of leukocytes into peripheral organs such as liver and kidneys (Fig. 2C and Supplementary Fig. 2, http://links.lww.com/HS/A96). The immunophenotype of the bone marrow cells was consistent for a myeloid leukemia that was positive for myeloid markers (Mac-1hi, Gr-1int) but not for lymphoid markers (CD3, B220) or progenitor markers (Sca-1, c-Kit) (Fig. 2D).
The presence of the reciprocal translocations t(9;17) in leukemia cells was confirmed by FISH using pan-chromosome probes (Fig. 2E). The Kmt2a-Mllt1 gene fusion and its product were verified by PCR and sequencing. Notably, both the non-rearranged allele of Kmt2a and Mllt1 as well as the reciprocal product Mllt1-Kmt2a fusion were also expressed in the leukemia cells (Fig. 2F). Transplantation of the bone marrow cells from the primary tumor mice into lethally irradiated second recipients manifested diseases in a shorter latency of 2–4 weeks, confirms the self-renewal capacity and the aggressiveness of this leukemia (Fig. 2B).
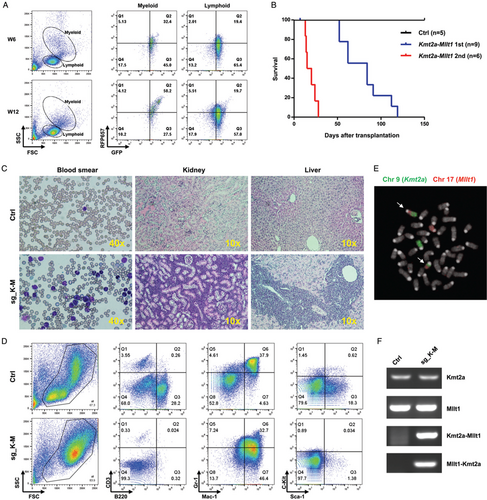
The Kmt2a-Mllt1 fusion induces acute myeloid leukemia. (A) Flow cytometry analysis of peripheral blood cells from a recipient mouse transplanted with Cas9GFP+/− bone marrow cells transduced with sg_K-M vectors at 6 weeks (W6) and 12 weeks (W12) after transplantation. A RFP657+ population with myeloid features is emerging over time. (B) Kaplan-Meier survival curves of mice transplanted with Cas9GFP+/− bone marrow cells transduced with sg_K-M vectors developing primary (1st) and secondary (2nd) Kmt2a-Mllt1 leukemia. (C) May-Grünwald Giemsa staining of blood smears and Hematoxylin-Eosin staining of tissue sections of kidneys and livers from control and Kmt2a-Mllt1 leukemia mice. (D) Fluorescence in situ hybridization (FISH) on metaphases of splenocytes from Kmt2a-Mllt1 leukemic mice using paints for chromosome 9 (FITC) or chromosome 17 (Cy3). White arrows indicate the translocated chromosomes. (E) Flow cytometry analysis of bone marrow cells stained with fluorescent antibodies in control and Kmt2a-Mllt1 leukemic mice. (F) RT-PCR detection of the expression of the non-translocated alleles of Kmt2a and Mllt1, and the fusions of Kmt2a-Mllt1 and Mllt1-Kmt2a in a control and a Kmt2a-Mllt1 leukemic mouse.
We next attempted to model the TCF3-HLF translocation defining a highly aggressive leukemia subtype for which so far no representative mouse model could be engineered.15 Given the frequent PAX5 heterozygous deletions and reduced gene expression identified in this leukemia,16 we tested the hypothesis if the Tcf3-Hlf translocation could induce a mouse leukemia when established in a Pax5 haploinsufficient context (Supplementary Fig. 3A–C, http://links.lww.com/HS/A96). We crossbred the Cas9 mice with the Pax5 heterozygous mice (Pax5+/−, C57BL/6 inbred)17 and used the F1 progenies (Cas9GFP+/− Pax5+/+ or Cas9GFP+/− Pax5+/−) as the bone marrow donors. Transduction of the bone marrow cells with validated sgRNAs targeting the equivalent introns of mice Tcf3 and Hlf loci (sg_T-H) lead to detectable Tcf3-Hlf translocation and the expression of the fusion product (Supplementary Fig. 3D-E, http://links.lww.com/HS/A96). The generation of the Tcf3-Hlf translocation was less effective than the Kmt2a-Mllt1 translocation, with ∼0.16% of the cells detected by ddPCR to harbor the translocation (Supplementary Fig. 3F, http://links.lww.com/HS/A96).
After transplantation of the engineered cells into lethally irradiated C57BL/6 recipients, the Tcf3-Hlf translocation was detected in genomic DNA from peripheral blood and bone marrow at 4 weeks after transplantation (Supplementary Fig. 3G-H, http://links.lww.com/HS/A96). However, no progression to leukemia could be detected even after a follow-up for up to 11 months. Post-mortem examination did not reveal any abnormality of spleen, kidney, liver and bone marrow cells and the Tcf3-Hlf translocation signal was undetectable in bone marrow cells (Supplementary Fig. 3H, http://links.lww.com/HS/A96). This suggests a selective disadvantage of cells carrying Tcf3-Hlf in vivo, confirming similar observations made with a knockin model of Tcf3-Hlf.18 Thus, despite the frequency of PAX5 haploinsufficiency in TCF3-HLF positive patients, deletion of Pax5 does not counteract the negative selection pressure by Tcf3-Hlf. Notably, the Kmt2a-Mllt1 myeloid leukemia can be established in the Pax5 +/− background with similar dynamics as in the Pax5 +/+ background confirming that a reduced Pax5 gene dosage does not influence the leukemogenic activity of Kmt2a-Mllt1 (Supplementary Fig. 4, http://links.lww.com/HS/A96).
Activation of oncogenes by fusion or juxtaposition to ectopic regulatory elements constitutes important cancer initiating events. Here, we established an efficient strategy using the CRISPR/Cas9 system to generate leukemogenic chromosomal translocations in vivo. We generated two types of translocations in bone marrow cells with frequencies of 0.6% and 0.16%. Although the generation of Tcf3-Hlf was not sufficient to induce leukemia in this study the model can now be used to evaluate different cells of origin and to screen for required cooperative genetic events. Our approach can be employed to model any pair of genes as long as the orientation to the centromere does not result in dicentric chromosomes. Using the inbred Cas9 mouse strain will further simplify this approach. Besides, our vector design facilitates the combination of multiple sgRNAs to introduce additional oncogenic lesions (Supplementary Fig. 1B, http://links.lww.com/HS/A96). Thus, our strategy provides an efficient way to generate precise translocations, enabling functional modeling in mice and constitutes a framework to study the role of cooperative events and the cells of origins in leukemogenesis.
Acknowledgments
We thank Meinrad Busslinger (Research Institute of Molecular Pathology, Vienna) for providing the Pax5+/− mice. We thank David Hediger (University Children's Hospital Zurich) for technical assistance. We also thank Raffaele Renella (University Hospital of Lausanne) and Dirk Heckl (Martin Luther University Halle-Wittenberg) for scientific discussions. This work was supported by the Swiss Cancer League (KFS-3526-08-2014; KFS-4237-08-2017) and the Swiss National Research Foundation SNF (310030-133108 and 323530-164223), the foundation ‘Kinderkrebsforschung Schweiz’, the ‘Krebsliga Zurich’, the Novartis Foundation of Biomedical Research, the clinical research focus program ‘Human Hemato-Lymphatic Diseases’ of the University of Zurich.