GTP Cyclohydrolase I Gene Expression in the Brains of Male and Female hph-1 Mice
Abbreviations used : BH4, 6R-l-erythro-5,6,7,8-tetrahydrobiopterin ; DA, dopamine ; DIG, digoxigenin ; GTPCH, GTP cyclohydrolase I ; HPD, hereditary progressive dystonia ; 5-HT, serotonin ; MA, monoamine ; MANOVA, multiple analysis of variance ; NE, norepinephrine ; SERT, serotonin transporter ; SRY, sex-determining Y region ; TH, tyrosine hydroxylase ; wt, wild type.
Abstract
Abstract : The hph-1 mouse is characterized by low levels of GTP cyclohydrolase I (GTPCH) and tetrahydrobiopterin. A quantitative double-lable in situ hybridization technique was used to examine CNS GTPCH mRNA expression within serotonin, dopamine, and norepinephrine neurons of male and female wild-type and hph-1 mice. In wild-type male and female animals the highest levels of GTPCH mRNA expression were observed within serotonin neurons, followed by norepinephrine and then dopamine neurons. Wild-type female animals were found to express lower levels of GTPCH mRNA in each cell type when compared with levels seen in wild-type males. GTPCH mRNA abundance in all three cell types was lower in hph-1 male than in wild-type male mice, with the greatest reduction in serotonin neurons. GTPCH mRNA levels were also lower in hph-1 female than in wild-type female mice, again with the greatest reduction occurring in serotonin neurons. Comparison of hph-1 male and hph-1 female mice revealed that the sex-linked difference in GTPCH mRNA expression observed in wild-type neurons was only present within female dopamine neurons. Overall, these results indicate that not only are basal levels of GTPCH mRNA expression heterogeneous across wild-type murine monoamine cell types but that gene expression is also modified in a sex-linked and cell-specific fashion by the hph-1 gene locus. The hph-1 mutation does not lie within the GT-PCH mRNA coding region. The 5′ flanking region of the GTPCH gene was cloned and sequenced and shown to be identical for both wild-type and hph-1 genomic DNA. Transient transfection assays performed in PC12 cells demonstrated that this 5′ flanking region was sufficient to initiate transcription of a luciferase reporter gene. Although the hph-1 mutation does not lie within the 5′ flanking region of the GTPCH gene, this region of the gene can function as a core promoter and is thus crucial to the control of GTPCH gene expression.
6R-l-5,6,7,8-Tetrahydrobiopterin [6-(R)-l-erythro-1′,2′-dihydroxypropyl)-2-amino-4-hydroxy-5,6,7,8-tetrahydropteridine ; BH4] is the required cofactor for the family of aromatic amino acid monooxygenases that includes tyrosine hydroxylase (TH), tryptophan hydroxylase, and phenylalanine hydroxylase (Kaufman, 1974). BH4 is also essential for the activity of the nitric oxide synthases (Marletta, 1993). GTP cyclohydrolase I (GTPCH) is the initial and rate-limiting enzyme in the BH4 biosynthetic pathway (Nichol et al., 1985). Humans homozygous for mutations in GTPCH exhibit phenylketonuria and low levels of CNS monoamine (MA) neurotransmitters (Blau et al., 1996). Ichinose et al. (1994) were the first to link heterozygous mutations in GTPCH with a movement disorder known as hereditary progressive dystonia (HPD) with marked diurnal fluctuation (Segawa et al., 1971, 1976), otherwise known as DOPA-responsive dystonia (Nygaard et al., 1991). HPD is a childhood-onset disease that is transmitted in an autosomal dominant manner with incomplete penetrance ; more female than male carriers are affected (Segawa et al., 1976 ; Nygaard et al., 1990, 1991). HPD is characterized by a decreased capacity to synthesize dopamine (DA) within intact nigrostriatal DA neurons (Rajput et al., 1994 ; Furukawa et al., 1995), and it is this imbalance within the neurochemical circuitry of the basal ganglia that presumably underlies the movement disorders associated with this disease. Although not overtly phenylketonuric, HPD patients also exhibit slow clearance of blood phenylalanine following a phenylalanine load (Hyland et al., 1997). Although the genetic basis for HPD has been largely established, these major phenotypic characteristics of HPD remain unexplained.
The hph-1 mutation was induced in mice by chemical mutagenesis in an attempt to produce an animal model of classical phenylketonuria (Bode et al., 1988). Subsequent work has shown that although these animals are mildly phenylketonuric, this mutation does not involve the phenylalanine hydroxylase gene locus but rather is associated with a large decline in GTPCH enzyme activity and the BH4 content of the liver (McDonald et al., 1988). Levels of BH4 within the brain are also significantly reduced by the hph-1 mutation (Hyland et al., 1996), albeit to a lesser degree than is found in liver (McDonald et al., 1988 ; Gutlich et al., 1994). Based on these phenotypic characteristics, the hph-1 genotype has been proposed as an animal model of HPD (Hyland et al., 1996).
Although the exact placement of the hph-1 locus within the mouse genome is unknown, the genotype has been mapped to chromosome 14 (Bode et al., 1988), on which is also found the gene for GTPCH (Ichinose et al., 1995). The coding region of hph-1 GTPCH mRNA is reported to be identical to that of the wild-type (wt) mRNA (Gutlich et al., 1994). Levels of GTPCH mRNA are, however, severely reduced in the hph-1 mouse liver, and this decrease presumably underlies the loss of GTPCH enzyme activity (Gutlich et al., 1994). The molecular basis for the reduction in content of GTPCH mRNA most likely involves mutation of cis- or trans-acting elements that control GTPCH gene transcription or mRNA stability.
In the present study, the effect of the hph-1 gene locus on the abundance of GTPCH mRNA within MA neurons in the brains of male and female mice was studied using the double-label in situ hybridization technique. In addition, the 5′ flanking region of the GTPCH gene was cloned and sequenced from both wt and hph-1 genomic DNA and shown to function as a core promoter within a heterologous promoter assay. These studies indicate a complex interplay among cell type, sex, and GTPCH gene expression within CNS MA neurons of both wt and hph-1 mice.
MATERIALS AND METHODS
Animals and tissue preparation
All animal handling procedures were performed according to the guidelines of the Baylor Institute Animal Care and Use Committee. Twelve hph-1 mice (six female and six male) maintained on the C57BL/6J × CBA.Ca background (Bode et al., 1988) and 12 C57BL/6J × CBA.Ca wt mice (six female and six male), all 2 months of age, were decapitated, and the brains were quickly removed and frozen in isopentane on dry ice. Serial sections 10 μm thick were taken through the substantia nigra zona compacta, dorsal raphe nucleus, and locus coeruleus of each brain. Sections were thaw-mounted onto gelatin-coated slides and kept desiccated at -80°C until use. Before being used for in situ hybridization, representative sections from each subject were stained with cresyl violet, and the anterior-posterior planes were matched across subjects.
Riboprobe synthesis
35S-CTP (1,250 Ci/mmol) was purchased from Du Pont New England Nuclear (Boston, MA, U.S.A.). A 35S-labeled 302-nucleotide cRNA probe specific to rat GTPCH cDNA and corresponding to nucleotides 269-570 was synthesized as previously described (Hirayama et al., 1993 ; Lentz and Kapatos, 1996). TH-positive neurons were identified using a digoxigenin (DIG)-labeled 282-nucleotide cRNA probe specific to rat TH nucleotides 1,240-1,521 (Hirayama et al., 1993). Serotonin (5-HT) neurons were identified using a DIG-labeled 155-nucleotide 5-HT transporter (SERT) cRNA corresponding to nucleotides 133-287 (Blakely et al., 1991 ; Burchett and Bannon, 1997). DIG-labeled cRNA probes were synthesized using the Boehringer-Mannheim (Indianapolis, IN, U.S.A.) Genius 4 RNA labeling kit.
In situ hybridization
In situ hybridization was performed according to the method of Lentz and Kapatos (1996) with minor modifications. Each individual in situ experiment consisted of matched sections of each brain region from two animals of each sex and genotype. Three independent in situ experiments were performed for a total of six animals per sex and genotype. Following prehybridization, 25 μl of hybridization solution (prehybridization solution containing 60 fmol of in vitro transcribed 35S-CTP-labeled GTPCH cRNA and either DIG-labeled TH or SERT cRNA) was applied to each section. Hybridization reactions were carried out at 52°C in a humid chamber for 4 h. Following hybridization the slides were incubated in blocking solution containing 10% fetal calf serum for 1.5 h at room temperature. Slides were next incubated for 12 h with anti-DIG antibody conjugated with alkaline phosphatase (1 : 2,000 dilution). Slides were then washed in washing buffer [100 mM Tris-HCl (pH 9.5), 100 mM NaCl, and 50 mM MgCl2] and incubated for 20 h in the dark at room temperature in color substrate solution (200 μl of nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate stock solution/10 ml of washing buffer). The color reaction was stopped by washing, and slides were airdried before autoradiography. Dried slides were dipped in Ilford K5 nuclear track emulsion (Polysciences, Warrington, PA, U.S.A.) diluted 1 : 1 with water and exposed at 4°C for 12 weeks. The slides were developed in Kodak D-19 developer at 18°C for 2 min, fixed with Kodak fixer for 4 min, rinsed in water for 15 min, and coverslipped with crystal mounting medium. Photomicrographs of the three monoaminergic cell groups were taken at both low- (31×) and high-power (1,260×) magnification under bright-field optics using a Zeiss Photomicroscopic III and Kodak color slide film (Tungsten 160T).
Quantitative image analysis
A Micro Computer Imaging Device (MCID/M2 ; Imaging Research, St. Catharines, Ontario, Canada) was used to quantify silver grains associated with TH- or SERT-positive neurons. Individual cells were magnified 1,260-fold under brightfield optics. Proportional area, the area of the cell occupied by silver grains, was determined as previously described (Lentz and Kapatos, 1996). Fifty cells per cell group were randomly chosen and analyzed from each of the 24 animals. For this analysis the experimenter was blind to both genotype and sex. Data obtained from ~3,600 individual neurons were then combined across experiments, and the three main variables of genotype, sex, and cell type were analyzed statistically by multiple ANOVA (MANOVA). Post hoc comparison with Tukey's test was used to test for significant differences between groups (p < 0.05).
Genomic DNA isolation and GTPCH gene sequencing
Frozen wt or hph-1 mouse brain was pulverized in liquid nitrogen, and genomic DNA was isolated. A set of primers was designed to generate an 896-bp product (-683 to +213) containing as much sequence 5′ to the translation start site as possible (Nomura et al., 1993 ; Ichinose et al., 1995). Primers used were CAGGTACCAGAGAGCAAGGTGTGTGT with a KpnI anchor for the upstream region and GAGCAGCGAGAGCAGAATGGAC for the downstream region. PCR reaction mix contained 1 ×Pfu buffer, 0.5 mM deoxynucleotide triphosphate mix, 1 μg of DNA template, each primer at 0.25 μM, 5% dimethyl sulfoxide, and 2.5 U of Pfu DNA polymerase. Template DNA was denatured at 95°C for 10 min before addition of Pfu. The PCR reaction was carried out at 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min for 35 cycles followed by 72°C for 10 min. PCR products were gel-purified, cloned into pGEM-7Zf (+) at SmaI and KpnI sites, and used to transform competent bacteria. Colonies were screened by PCR, and those found to be positive for GTPCH genomic sequence were amplified.
DNA sequencing
Wt and hph-1 GTPCH DNAs (718 bp) were both amplified as described above except that a downstream internal primer, GGAGTCAGGTGCACCAATGG, corresponding to nucleotides +16 to +35 was used. PCR products were cloned into pGEM3Z (+), and both strands of DNA were sequenced using SP6 or T7 primers.
Promoter activity assay
To produce p0.681 GTPCHluc, 681 bp of the wt proximal promoter up to and including the GTPCH translation start site was cloned into the KpnI/NcoI sites of the pGL3-basic plasmid vector. Transient transfection of PC12 cells plated in 24-well plates was performed using Lipofectamine (GibcoBRL) and a combination of 0.4 μg of p0.681GTPCHluc and 0.1 μg of pRSVβ-galactosidase DNA. Promoterless pGL3-basic and SV40 promoter-driven pGL3-control plasmid DNA served as control and reference transfections, respectively. All transfections were done in triplicate, and 48 h later cell lysates were assayed for luciferase and β-galactosidase using chemiluminescence substrates. Differences in transfection efficiency across wells were normalized by β-galactosidase activity, and luciferase activity was expressed as relative light units.
RESULTS
In the rat brain GTPCH mRNA detected by in situ hybridization is localized exclusively to MA neurons (Hirayama et al., 1993 ; Lentz and Kapatos, 1996). Nonetheless, it seemed prudent not to make this same assumption in the case of the hph-1 mouse brain. The doublelabel in situ hybridization technique was therefore used to colocalize GTPCH mRNA with the mRNAs encoding for TH or SERT, transcripts that are phenotypic markers for DA and norepinephrine (NE) or 5-HT neurons, respectively. In these experiments, the purple alkaline phosphatase reaction product associated with DIG-labeled TH or SERT cRNA was used to identify cell type, whereas simultaneous autoradiography with a 35S-labeled GTPCH cRNA was used to detect and quantify GTPCH mRNA. Figure 1 contains low-power photomicrographs of the three wt MA cell groups from a typical double-label in situ hybridization experiment. The DA neurons within the substantia nigra (Fig. 1A), 5-HT neurons of the dorsal raphe (Fig. 1B), and NE neurons within the locus coeruleus (Fig. 1C) can each be easily identified by the purple chromogen as containing TH or SERT mRNA. No obvious difference in TH or SERT DIG-labeling intensity or in the number of labeled neurons was observed in brain sections of male and female wt or hph-1 mice. Autoradiographic grains indicative of GTPCH mRNA were observed under high-power magnification to be associated with the purple chromogen of TH mRNA-containing DA neurons of the substantia nigra (Fig. 1D) and NE neurons of the locus coeruleus (Fig. 1F) or SERT mRNA containing 5-HT neurons of the dorsal raphe (Fig. 1E). Regardless of sex or genotype, autoradiographic grains were only associated with the DIG-labeled neurons of each MA cell group. Moreover, in each cell group the number of grains appeared to be substantially lower within hph-1 DA (Fig. 1G), 5-HT (Fig. 1H), and NE (Fig. 1I) neurons.
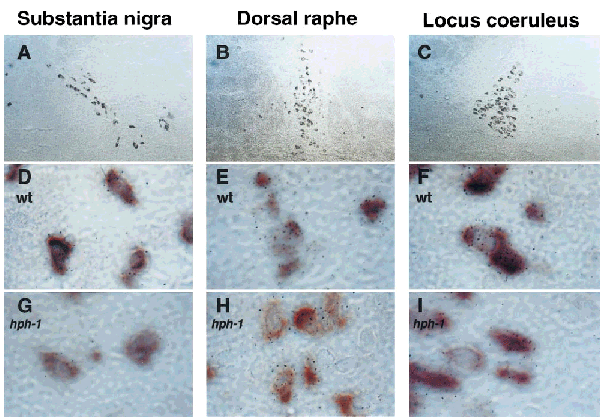
Double-label in situ hybridization for GTPCH, TH, and SERT mRNA. Bright-field low-power (31× ; A-C) and high-power (1,260 × ; D-I) photomicrographs were obtained of wt and hph-1 MA neurons. DA (A, D, and G) and NE (C, F, and I) neurons of the substantia nigra and locus coeruleus were identified by the purple DIG-TH mRNA reaction product. 5-HT neurons of the dorsal raphe (B, E, and H) were identified by the purple DIG-SERT mRNA reaction product. GTPCH mRNA was identified by the presence of autoradiographic grains.
Effect of hph-1 mutation on GTPCH mRNA expression in MA neurons
Autoradiographic grain densities associated with each identified cell type were determined using quantitative image analysis and were expressed as proportional areas. MANOVA revealed significant main effects of genotype, cell type, and sex as well as significant interactions between genotype and cell type. Figure 2A illustrates a comparison of GTPCH mRNA expression across 5-HT, DA, and NE cell types of wt animals that have been grouped according to sex. In both males and females a hierarchy in GTPCH mRNA expression across MA cell groups was observed (F2,1744 = 124.5 ; p < 0.001), with significantly higher levels measured within 5-HT followed by NE and then DA neurons. Overall, when compared with their male counterparts, female wt animals were found to express lower levels of GTPCH mRNA (F1,1744 = 47.6 ; p < 0.001), and this 20-35% difference was significant for each cell group. Based on these observations, all further data from female and male subjects were analyzed separately to determine the effect of the hph-1 locus on GTPCH mRNA levels.
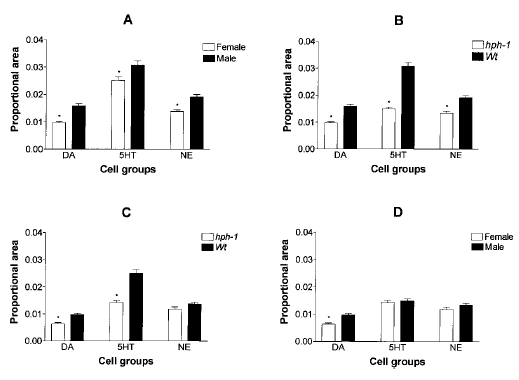
Quantification of GTPCH mRNA expression in hph-1 and wt mice. GTPCH mRNA abundance was expressed as proportional area. A : GTPCH mRNA expression in male and female wt mice. B : GTPCH mRNA expression in male wt and hph-1 mice. C : GTPCH mRNA expression in female wt and hph-1 mice. D : GTPCH mRNA expression in male and female hph-1 mice.
Figure 2B shows that GTPCH mRNA abundance across all three cell groups was significantly lower in male hph-1 than in male wt mice (F1,1794 = 160.8 ; p < 0.001). MANOVA also showed an interaction between genotype and cell type (F2,1494 = 20.9 ; p < 0.001), which reflects a significantly greater reduction in 5-HT (51%) than in DA (39%) or NE (30%) neurons. Owing to the large reduction in transcript levels within 5-HT neurons, the hierarchy observed in GTPCH mRNA expression in wt animals was disrupted in hph-1 animals. In comparison with 5-HT and NE neurons, however, significantly lower levels of expression were maintained within hph-1 DA neurons.
Figure 2C shows that GTPCH mRNA levels are decreased significantly in female hph-1 mice relative to female wt mice (F2,1494 = 58.2 ; p < 0.001). MANOVA again detected a significant interaction between genotype and cell type (F2,1494 = 15.9 ; p < 0.001), which reflects a greater reduction in 5-HT (43%) than in DA (35%) neurons. The 14% decrease in GTPCH mRNA content in female hph-1 NE neurons did not reach statistical significance.
Figure 2D is a comparison of GTPCH mRNA abundance in male and female hph-1 mice. Although MANOVA showed that transcript levels in male and female hph-1 mice are different with respect to sex (F1,1544 = 11.5 ; p < 0.001) and cell type (F2,1544 = 55.1 ; p < 0.001), the post hoc analysis indicated that GTPCH mRNA is maintained at significantly lower levels only in female hph-1 DA neurons.
DNA sequencing and functional analysis of wt and hph-1 GTPCH core promoter region
The molecular basis for the reduction in GTPCH mRNA content in MA neurons of the hph-1 mouse may involve mutation of cis-acting elements that are important for the control of GTPCH gene transcription. The GTPCH 5′ flanking region corresponding to nucleotides -1 through -681 were therefore cloned and sequenced from wt and hph-1 genomic DNA. A comparison of wt and hph-1 sequences did not detect a single alteration in base composition within this 681 bp (Fig. 3). With the exception of a few base changes this sequence is identical to that reported previously for a mouse of unknown genotype (Ichinose et al., 1994). Further analysis did identify, however, several putative cis-acting elements that may be involved in assembly of the basal transcription machinery, including a TATA box at bp - 175 to - 169 (ATAAAAA), a CAAT box at bp -216 through -212 (CCAAT), a partial CRE at bp -234 to -227 (TGACGCAA), and SPl sites at bp -264 through -255 (GGGCGGGGCT) and -252 through -244 (GGGGCGGGGA). It is interesting that three putative sex-determining Y region (SRY) elements were detected at -299 through -305 (CTTGTTT), -393 through -399 (GGTGTTT), and -365 through -371 (AAAGAAA).

DNA sequence of wt and hph-1 GTPCH 5′ flanking regions. Genomic DNA was isolated from wt and hph-1 brains and used as the template in PCR reactions. The top sequence in bold letters is from the wt DNA, and the bottom sequence with italic letters is from hph-1 DNA. Putative cis-acting elements are boxed and labeled.
Studies using transient transfections assays were next undertaken to demonstrate that the cloned 5′ flanking region of GTPCH can function as a core promoter to initiate transcription of the heterologous luciferase reporter gene. PC12 cells were chosen for this analysis because they constitutively express GTPCH. Normalized luciferase activity of PC12 cells transfected with p0.681 wt GTPCHluc, pGL3-basic, or pGL3-control DNA is presented in Fig. 4. Extracts of PC12 cells transfected with p0.681wtGTPCHluc DNA were found to contain >20 times the luciferase activity of the promoterless pGL3-control and approximately one-half of the activity generated by the strong viral promoter-driven pGL3-control vector.
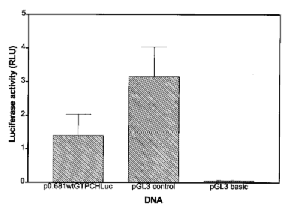
Transient transfection assay of GTPCH promoter activity in PC12 cells. All transfections were done in triplicate, and 48 h later cell lysates were assayed for luciferase and β-galactosidase. Differences in transfection efficiency across wells were normalized by β-galactosidase activity, and luciferase activity was then expressed as relative light units (RLU).
DISCUSSION
We have studied GTPCH mRNA expression in 5-HT, NE, and DA neurons from male and female mice of both the wt and hph-1 genotypes. Several important conclusions can be reached from these data, some of which may be relevant to our understanding of HPD.
There are cell type-dependent differences in GTPCH mRNA abundance in both wt and hph-1 mouse brain
In both male and female wt mice the 5-HT neurons of the dorsal raphe were found to contain significantly more GTPCH mRNA than the NE neurons of the locus coeruleus, whereas the DA neurons of the substantia nigra contained significantly less than either 5-HT or NE cell types. These results are in complete agreement with those reported previously for male rat brain (Lentz and Kapatos, 1996). Decreases in GTPCH mRNA content in each cell group of hph-1 mice were detected and, depending on the sex of the animal, ranged from a high of 51% in 5-HT neurons of the male to a low of 14% in NE neurons of the female. Overall, the decline in GTPCH abundance in male and female hph-1 mice was significantly greater in 5-HT neurons than in the other two cell populations. The cell type-dependent hierarchy in GTPCH mRNA expression observed in wt animals continued to be partially maintained in hph-1 mice of either sex, in that hph-1 DA neurons were found to maintain the lowest abundance of GTPCH mRNA.
There are sex-linked differences in GTPCH mRNA abundance in both wt and hph-1 mouse brain
5-HT, NE, and DA neurons of female wt animals were each found to contain less GTPCH mRNA than those of their wt male counterparts. Nonetheless, when compared with the wt expression pattern, there was a larger decline in GTPCH mRNA content for all three cell groups detected in hph-1 males when compared with hph-1 females. Despite the larger decline in male hph-1 mice, GTPCH abundance in female hph-1 DA neurons remained significantly lower when compared with hph-1 males.
The hph-1 mutation does not lie within the GTPCH proximal promoter
Levels of GTPCH mRNA in the hph-1 liver (Gutlich et al., 1994) and brain (this study) are decreased despite the fact that the GTPCH core promoter (this study) and mRNA (Gutlich et al., 1994) are identical to the wt. Ichinose et al. (1995) have also reported on a patient with HPD and low GTPCH enzyme activity but no mutation in the GTPCH coding region. These observations raise the possibility that mutations in important trans- or cis- acting elements may be involved in decreasing GTPCH gene expression in the hph-1 mouse and in some cases of HPD. This idea finds support in reports of developmental increases in liver GTPCH enzyme activity and BH4 content in hph-1 animals (McDonald et al., 1988 ; Gutlich et al., 1994 ; Hyland et al., 1996) and in the disparate effect reported here of the hph-1 gene locus on GTPCH mRNA expression within CNS MA neurons. In an attempt to discover a mutation in the 5′ flanking region of the hph-1 GTPCH gene that is capable of decreasing the rate of basal transcription, we have cloned 681 bp of wt and hph-1 genomic DNA. Sequence analysis revealed, however, that the wt and hph-1 mouse genomes are identical in this region. These results do not rule out the possibility that the hph-1 mutation may be found further upstream of the region sequenced here or within one of the five GTPCH gene introns (Ichinose et al., 1994).
Several putative cis-acting elements were detected in this 5′ flanking region, perhaps the most interesting of which includes three copies of the SRY element (Pontiggia et al., 1995). The gene coding for the transcription factor SRY is found on the Y chromosome and as such is expressed exclusively in male animals. SRY contains the high-mobility group motif and is detected in only the testis and the brain. It therefore seems possible that SRY or some related transcription factor may be involved in the sex-linked differences in GTPCH gene expression reported here. Transient transfection assays demonstrated that the 681-bp 5′ flanking region of the GTPCH gene does contain the requisite cis-acting elements to initiate and maintain transcription of the luciferase gene. Although the hph-1 mutation does not lie within this 5′ flanking region, this region of the gene can function as a core promoter and is thus crucial to the control of GTPCH gene expression. It is highly likely that mutations in this core promoter will be found capable of decreasing GTPCH gene transcription and may be of clinical importance.
Heterogeneity in GTPCH gene expression in the brain has important implications for determining steady-state levels of BH4 and the regulation of MA biosynthesis (Lentz and Kapatos, 1996). BH4 levels have been shown to be a limiting factor for TH activity in DA neurons (Kettler et al., 1974 ; Miwa et al., 1985) but not for tryptophan hydroxylase activity in 5-HT neurons (Wolf et al., 1990, 1991). Nonetheless, the magnitude of the decrease in GTPCH mRNA expression within 5-HT neurons presumably underlies the greater decrements in levels of 5-HT and metabolites found in the brain of hph-1 animals (Hyland et al., 1996). In some children with inborn errors of BH4 metabolism, levels of CSF 5-HT metabolites are more severely affected than are those of metabolites of DA or NE (Dhondt, 1991). These same children, however, exhibit no obvious physiological deficits resulting from decreased 5-HT synthesis yet do show disorders of movement that are indicative of low levels of DA within the basal ganglia (Kaufman et al., 1983). Neurotransmitter synthesis within human nigrostriatal DA neurons may therefore be selectively vulnerable to deficits in BH4 biosynthesis. As shown here and in earlier studies (Lentz and Kapatos, 1996), in comparison with 5-HT and NE neurons, nigrostriatal DA neurons contain low levels of GTPCH mRNA. In addition, recent work has also demonstrated that the amount of GTPCH protein within nigrostriatal DA neurons is even lower than would be predicted based on the abundance of GTPCH mRNA (Hirayama and Kapatos, 1998). Mutations in GTPCH may therefore interact with the already low level of GTPCH gene expression within the nigrostriatal DA neurons to drive BH4 levels below a minimal threshold level required to maintain DA synthesis. Moreover, sex-linked difference in GTPCH mRNA expression within nigrostriatal DA neurons may predispose female carriers to the HPD phenotype.
Overall, these results indicate that not only are basal levels of GTPCH mRNA expression heterogeneous across murine 5-HT, NE, and DA cell types, but that gene expression is also modified in a sex-linked and cell-specific fashion by the hph-1 gene locus. Several intriguing questions concerning GTPCH mRNA expression are raised by these data. Why do wt female animals express lower levels than wt males ? Why do male hph-1 animals appear to be more affected than females ? Why are 5-HT neurons of either sex most affected by the hph-1 mutation ? Why is the male-female difference in DA but not 5-HT or NE neurons maintained in the hph-1 genotype ? Because there are several obvious parallels between the results presented here and what is known about HPD, answers to these questions may shed some light on the biological basis for this neurological disorder.
Acknowledgements
We thank Dr. M. Bannon and Mr. S. Burchett for the generous gift of SERT probe template, Dr. C. Whitty for assistance in quantitative image analysis, and Dr. R.-Y. Shen for statistical consultation. This work was supported by the Dystonia Medical Research Foundation and grant NS26081 (to G.K.) from the National Institutes of Health.




