A fixed allele at microsatellite locus LS-39 exhibiting species-specificity for the black caviar producer Acipenser stellatus
Abstract
The sturgeon microsatellite LS-39 was amplified across 10 different species of Acipenserinae and exhibited the potential to identify the black caviar producer Acipenser stellatus on a genomic DNA level. This was because of a fixed allele of 111 bp, which was absent in the other species which were investigated. Concerning the source of sturgeon species, LS-39 is the first nuclear marker described to examine black caviar. Furthermore, new light is shed on the controversial ploidy state of sturgeons. The present authors findings at this microsatellite locus support the hypothesis that extant ~120 chromosomal species are modern diploids, whereas sturgeons with ~240 chromosomes should be considered as modern tetraploids.
Introduction
Almost all Acipenseriformes species are threatened and some species are near extinction (Birstein et al. 1997a). The dramatic decrease of natural populations is attributed to anthropogenic influences, i.e. hydraulic engineering, pollution and massive overfishing for black caviar (Birstein 1993; Bemis and Findeis 1994). Traditionally, the three sturgeon species, Huso huso L., Acipenser gueldenstaedtii Brandt, and Acipenser stellatus Pallas, are used commercially for caviar production. However, in recent literature, it has been reported that mislabelled, highly prized caviar lots were found on the international market (DeSalle and Birstein 1996; Birstein et al. 1998b), indicating the need to protect consumers, wholesalers and the toll against deceit. Moreover, identifying the origin of the caviar may also prevent the exploitation of endangered, non-commercial sturgeon species (DeSalle and Birstein 1996).
To date, the origin identification of black caviar has been limited to the use of mitochondrial DNA markers (DeSalle and Birstein 1996; Birstein et al. 1998a, b). Attempts to distinguish among sturgeon species on the genomic DNA level have been unsatisfactory (Genetic Analyses Inc. 1994; Birstein et al. 1997b).
In contrast to other fish species, such as Atlantic salmon (Slettan et al. 1997), Nile tilapia (Kocher et al. 1998) or zebrafish (Shimoda et al. 1999), only 11 microsatellite loci have been characterized for A. fulvescens Rafinesque (May et al. 1997). These co-dominant genomic markers are inherited in Mendelian fashion and have been proven to cross-amplify among the genera Acipenser and Scaphirhynchus (May et al. 1997). Apart from potential genetic population studies, microsatellites are also suitable to elucidate the ploidy level of acipenseriform species, an approach which is still controversial. Some authors have suggested that sturgeons with ~120 and ~240 chromosomes are tetraploid and octaploid, respectively (for review, see Birstein et al. 1997b), whereas Fontana (1994), for example, described these as di- and tetraploid, respectively.
The present study focused on the sturgeon microsatellite marker LS-39 and its suitability in the identification of sturgeon species. Moreover, its use in determining ploidy levels is also indicated.
Materials and methods
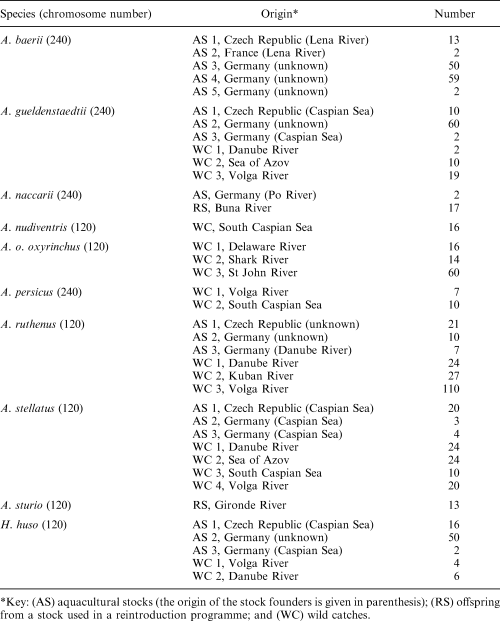
Blood and fin clips, as well as roe samples of 102 A. gueldenstaedtii, 105 A. stellatus, 78 H. huso, 126 A. baerii Brandt, 19 A. naccarii Bonaparte, 16 A nudiventris Lovetzky, 203 A. ruthenus L., 17 A. persicus Borodin, 13 A. sturio L. and 90 A. oxyrinchus oxyrinchus Mitchill were used for DNA extractions according to the QIAGEN blood and tissue protocols (QIAGEN Inc., Hilden, Germany). Detailed information on the origin and number of samples, as well as the species’ chromosomal state are given in Table 1.
The primer sequences for LS-39 are 5′-TTCTGAAGTTCACACATTG-3′ (LS-39F, forward primer) and 5′-ATGGAGÍCATTATTGGAAGG-3′ (LS-39R, reverse primer) (May et al. 1997). Primer LS-39F was fluorescently labelled with 6-FAM (PE Applied Biosystems, Weiterstadt, Germany). Amplifications were performed in 25-μl volumes containing 50 ng genomic DNA, 0·25 U Taq polymerase (MBI-Fermentas, Vilnius, Lithuania), 5 picomoles of each primer, 0·10 mM Tris-HCl (pH 8·8 at 25°C), 50 mM KCl, 1·5 mM MgCl2, 0·1 μg μl−1 bovine serum albumin (BSA), 0·08% (v/v) Nonidet P40 and 100 μM of each dNTP. Amplification was accomplished with 30 cycles in the following steps: 30 s at 94°C, 30 s at 57°C, 30 s at 72°C and a 5-min final extension at 72°C. The sizes of the alleles were determined on a DNA sequencing automate (Model 310, PE Applied Biosystems) using the software package 310 GeneScan® 2.1.
Results
Gene dosage derivation
For species with ~120 chromosomes, one or two peaks were detected in individual electropherograms at microsatellite locus LS-39. In contrast, specimens of the remaining sturgeon taxa with ~240 chromosomes displayed a maximum of four peaks differing in size. While a single peak observed in the electropherogram of a specimen was considered to represent four alleles of the same size, four detected peaks differing in size were considered to represent four different alleles. For those fish exhibiting two or three peaks differing in size, the ratio of peak heights and areas was used in deriving gene dosages. An example is given in Fig. 1. In the first fish (Fig. 1a), two peaks were detected in the electropherogram. The height (area) of the 120-bp peak was approximately threefold larger than the height (area) of the peak observed at 123 bp (see Fig. 1c). Therefore, the 120-bp peak was considered to comprise three alleles of this size, whereas only one allele contributed to the peak located at 123 bp. If the same derivation is applied to the second specimen (Figs 1b, c), both peaks observed at 117 and 123 bp, respectively, are represented by only one allele each, whereas the peak at 120 bp comprised two alleles.
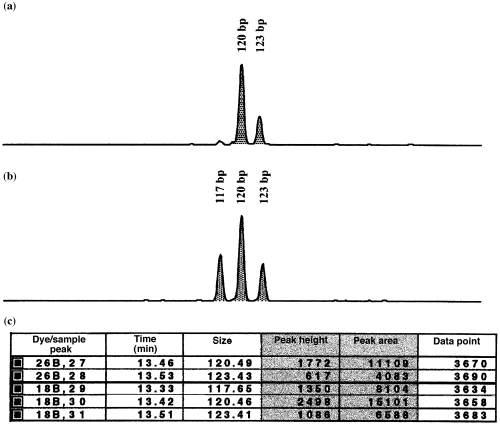
Examples of the electropherograms and peak data of microsatellite locus LS-39 in two Acipenser baerii individuals, as reícorded by the sequencing automate ABI PRISM 310 and using the data obtained in gene dosage derivation: (a) two peaks at 120 and 123 bp are observed in the electropherogram of fish 1 (see the first two rows in the ‘Size’ column in Fig. 1c); (b) three peaks detected at 117, 120 and 123 bp in the electropherogram of fish 2 (see the last three sample rows in the ‘Size’ column in Fig. 1c); and (c) sample information recorded by 310 GeneScan® 2.1 software. Within individuals, gene dosages are obtained by determining relative peak heights/areas. For fish 1, the 120-bp peak comprises three alleles and the peak located at 123 bp represents one allele. In fish 2, the relative values of 117:120:123-bp peaks are ~1:~2:~1
In some specimens of A. baerii and A. gueldenstaedtii, only three peaks of equal heights and areas were observed.
Allele sizes and frequencies
In total, 17 different alleles ranging between 108 and 156 bp were observed at locus LS-39. In contrast to the other sturgeon species examined, A. stellatus was fixed for the allele at 111 bp, which was absent in all other species studied. The highest number of alleles at this locus was observed in A. gueldenstaedtii. Because of the large number of populations analysed and the outcome of the present study concerning the species-specificity of LS-39, allele sizes and frequencies were hence presented as data which were pooled among populations within species (Table 2).
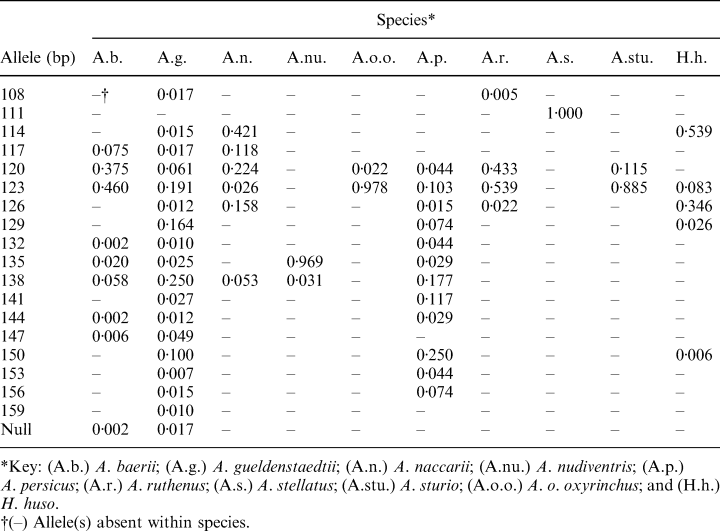
Because of the tetraploidy of N 240 chromosomal sturgeon species, neither heterozygosity data nor Hardy–Weinberg deviations were calculated.
Discussion
The use of mitochondrial genes in identifying sturgeon species has been described elsewhere (DeSalle and Birstein 1996; Ong et al. 1996; Wirgin et al. 1997; Birstein et al. 1998a,b; Ludwig and Kirschbaum 1998). However, natural as well as artificial hybridizations between different sturgeon species are known (for overview, see Birstein et al. 1997b), and hybrids between species with the same chromosome number are fertile (Nikoljukin 1966; Ronyai and Varadi 1995). Therefore, results which are merely based on the information of mitochondrial DNA markers should be considered with reservation. For instance, caviar derived from females of the so-called hybrid ‘bester’, which is produced by the cross of a H. huso female and an A. ruthenus male, would be identified as beluga caviar from H. huso solely on the basis of mitochondrial DNA markers. Thus, a reliable identification of black caviar species origin must include additional genomic markers. LS-39 is the first genomic marker described in sturgeons with a potential to identify the black caviar producer A. stellatus. This was because of a fixation of the allele occurring at 111 bp in each specimen of A. stellatus from every important caviar-producing population; this allele was absent in all other sturgeon taxa analysed. However, the circumstances concerning the presence of this allele solely in A. stellatus and its fixation in this species remain speculative. Interestingly, all taxa exhibiting ~120 chromosomes merely showed a quite low polymorphism at locus LS-39. In contrast to these findings, the ~240 chromosomal species A. baerii, A. gueldenstaedtii and A. persicus, in particular, were rather polymorphic at this locus. It might be assumed that constraints may act on LS-39 or another locus linked to LS-39, and thus, may have an impact on the variability at LS-39, especially in lower chromosomal sturgeon species. In contrast to taxa with ~120 chromosomes, ~240 chromosomal species have twice as many copies of a considered locus. Therefore, an overall reduced constraint may be assumed for species with the higher chromosome number, resulting in higher polymorphisms as observed at LS-39.
In addition to its species-specificity, microsatellite locus LS-39 also enabled partial insight into sturgeon cytogenetics. So far, controversial discussions are found in the literature concerning the ploidy level of different acipenseriform species. For instance, Fontana (1994) suggested that sturgeon taxa with ~120 chromosomes should be considered as diploids, whereas species with ~240 chromosomes were tetraploid. In contrast, Birstein et al. (1997b) quoted species with ~120 chromosomes as tetraploid, with those exhibiting ~240 chromosomes as octaploids. As presented in Table 1, the present study investigated taxa from both chromosomal groups. Because microsatellite loci are co-dominant markers, the data recorded at LS-39 is transferable into genotypic information for each specimen. Within a diploid species, a heterozygous individual has two alleles differing in size at a considered locus. In the present study, no individual among the ~120 chromosomal species was observed to have more than two alleles. Based on their results obtained at microsatellite locus LS-39, the present authors conclude that ~120 chromosomal sturgeon taxa are modern diploids. This is in accordance with Fontana (1994), but refutes the suggestion of Birstein et al. (1997b). On the assumption that sturgeon species with ~240 chromosomes are modern tetraploids, a maximum of four different alleles is expected to occur within specimens at a locus. In contrast to tetraploidy, more than four different alleles should be observed, assuming an octaploid genomic level as proposed by Birstein et al. (1997b). However, the present authors detected no more than four different alleles among specimens with ~240 chromosomes. With regard to the ploidy level of these species, the present data again provide evidence for the hypothesis postulated by Fontana (1994).
Surprisingly, some specimens of both the A. baerii and A. gueldenstaedtii taxa showed only three peaks of equal size and height. Such a triploidic peak pattern could be assumed to point to hybrids resulting from crosses between parental species differing in chromosome number, i.e. ~120 × ~240 or vice versa. However, because no one individual was identified as such a potential morphological hybrid and since these hybrids are assumed to be sterile (Nikoljukin 1966), the fourth allele was thus considered to be a null allele. On the one hand, the occurrence of null alleles has no impact on the suitability of microsatellite locus LS-39 in identifying A. stellatus. On the other, prior to an application of this microsatellite locus in population genetic studies of sturgeon species, a further investigation of the primer flanking sequences seems to be necessary. Because of the limited sample size of some tetraploid species, especially for A. naccarii and A. persicus, the occurrence of null alleles could not be excluded. In contrast to tetraploid sturgeons, null alleles are only detected in segregation analyses. Because of the lack of families in the present study, no conclusions concerning the occurrence of null alleles in ~120 chromosomal taxa could be drawn.
The results obtained in the present study indicate that some microsatellites may become valuable tools in sturgeon species and black caviar identification. Furthermore, this class of markers is sufficiently consistent to provide information concerning the ploidy state of these so-called ‘living fossils’.
Acknowledgements
We would like to thank C. Kaltwasser and J. Becker for their technical assistance, as well as J. Gessner, P. Willot, O. Goeímann, F. Kirschbaum, N. Patriche and T. Gulyas for providing materials. This work was supported by grants from the DFG (ME 712/12) and from the BfN by order of the BMU (FKZ: 808 05 078).