Phylogeography of the yucca moth Tegeticula maculata: the role of historical biogeography in reconciling high genetic structure with limited speciation
Abstract
Tegeticula maculata is one of the most ancient and morphologically variable lineages within the yucca moths, yet has apparently undergone little diversification in comparison with much younger yucca moth lineages that have rapidly diversified. A phylogeographic approach was used to determine the number of independent lineages within T. maculata and to examine whether these patterns corresponded with morphological differences between its subspecies maculata and extranea. Phylogenetic analysis of mitochondrial DNA sequence variation indicated that the two subspecies are in separate clades, but there was also an equally deep split within subspecies maculata. There was no evidence for gene flow among regions and there was considerable substructure within clades. The phylogeographic structure of moth populations among and within subspecies can be explained in part by historical biogeographic boundaries and increasingly patchy postglacial distribution of the exclusive host plant, Hesperoyucca whipplei. Local specialization and co-adaptation would be possible in the absence of apparent gene flow, yet gross morphological divergence is limited to the very old split between the subspecies. Sorting of ancient mitochondrial lineages followed by local genetic differentiation may explain the pattern of high genetic structure with limited speciation.
Introduction
The obligate mutualism between yucca moths and yuccas is widely considered one of the classic examples of co-evolution and it provides an excellent model system to examine the links between phylogeographic structure, emerging host specificity and patterns of speciation. Yucca moths serve as the exclusive pollinators of their hosts and their larvae in turn require some of the yucca seeds for their development (Riley 1892; Trelease 1893). Most of the 16 described moth species feed on a single host species, but some use as many as seven species across their geographic range (Pellmyr 1999). Differences in moth morphology, phenology and behaviour help delineate species and there is generally little intraspecific variation in these traits (Miles 1983; Powell 1992; Pellmyr 1999).
In contrast to other yucca moth species, Tegeticula maculata displays striking differentiation in wing pattern across its range (Fig. 1; Busck 1947; Powell & Mackie 1966; Davis 1967). Despite the remarkable difference in wing pattern, however, there is no recorded variation in genitalia between the T. maculata morphs, whereas the genitalia are highly variable and diagnostic among all congeners (Davis 1967; Pellmyr 1999). Originally described as three separate species (T. maculata, T. apicella and T. extranea;Edwards 1888; Riley 1892; Dyar 1903; Meyrick 1913; McDunnough 1939; Busck 1947), the morphs are considered intraspecific entities in the most recent revisions (Powell & Mackie 1966; Davis 1967). Powell and Mackie recognized all three as subspecies, whereas Davis relegated apicella into synonymy with maculata. We follow the latter arrangement here.
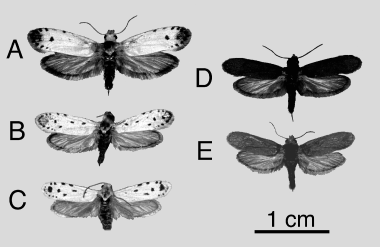
Wing pattern variation in Tegeticula maculata. (A) T. m. maculata (La Tuna Canyon, CA). (B) T. m. maculata (Kaweah, CA). (C) T. m. maculata (Kern Canyon, CA). (D) T. m. extranea (Pinyon Flat, CA). (E) T. m. extranea (Jaraguay Pass, Baja California, MX).
Both moth subspecies exclusively pollinate Hesperoyucca whipplei, the most ancient yucca pollinated by yucca moths (Bogler et al. 1995). Host plant populations are patchily distributed in coastal scrub and chaparral communities in southern California and northwestern Arizona, USA and in north-central Baja California, Mexico (Fig. 2; McKelvey 1947; Benson & Darrow 1981; Turner et al. 1995). The northern coastal and Sierra Nevada H. whipplei populations host the white speckled T. m. maculata, while the black T. m. extranea occurs in southern coastal California and Baja California populations (Fig. 2). The moth subspecies may come into contact in the San Gabriel Mountains north of Los Angeles, but speculations of introgression are limited to a single extranea individual with light scales on the cranial hindmargin (Davis 1967; O. Pellmyr, unpublished).
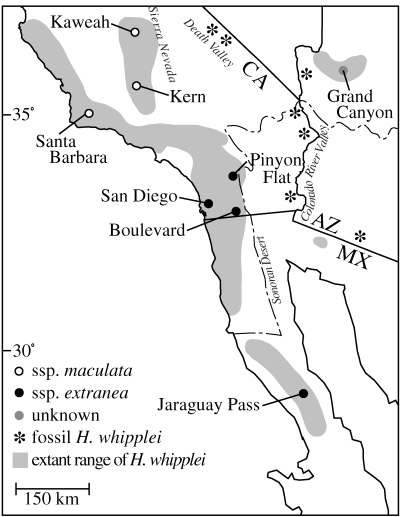
Distribution of collection sites for Tegeticula maculata. Open circles are collection sites of T. m. maculata (white speckled moths), black circles are collection sites of T. m. extranea (black moths) and the grey circle represents the Grand Canyon collection site with unknown phenotypic affinity. Shaded regions indicate the distribution of the host plant, Hesperoyucca whipplei, following Haines (1941) and McKelvey (1947). Asterisks denote Pleistocene and recent locations of H. whipplei following Wells & Woodcock (1985), Cole (1986), Woodcock (1986) and Van Devender (1987). The dotted line indicates the western boundary of the Sonoran desert.
If morphology is indicative of phylogenetic subdivision within T. maculata, moths with distinct phenotypic affinities should occur as discrete lineages in a phylogeographic analysis. We may also expect to find other divisions within T. maculata because it is an ancient lineage that arose near the initial radiation of the pollinator species about 40 Ma and surveys of younger yucca moth lineages have revealed explosive species radiations over much shorter time spans (Pellmyr & Leebens-Mack 1999). Hence, we predict additional substructure within each of the subspecies that corresponds to geographic variation in wing pattern. For example, moths from Jaraguay Pass in Baja California are smaller than typical T. m. extranea and differ in having light ventral scaling (Fig. 1). Structure within the subspecies may also be facilitated by periodic changes in the historical distribution of host plant populations that may have been important in structuring past gene flow among T. maculata populations. Fossilized packrat midden records indicate that H. whipplei spanned large regions of the Mojave and Sonoran deserts as recently as during the latest glacial (Spaulding 1990; Van Devender 1990) and its range has fragmented over at least the past 11 000 years. Repeated host range contraction–expansion events during the Pleistocene may have had considerable impact on the population dynamics of T. maculata.
The purpose of this study was to determine whether phylogeographic patterns within T. maculata support the proposed hypotheses of subspecies designations and of further structure among populations. Using estimates of relatedness among T. maculata populations derived from cytochrome oxidase I (COI) mitochondrial DNA (mtDNA) sequences, we addressed two specific questions. First, does the phylogeographic structure of moth populations reflect patterns of phenotypic variation, that is, are the named subspecies of T. maculata monophyletic, cohesive groups? Second, is there further structure within these subspecies that might mediate diversification, or does extensive gene flow among populations within recognized lineages counter diversification?
Materials and methods
Moths were collected from eight sites, spanning the extant geographic range of Tegeticula maculata (Fig. 2). Six individuals were sampled from each population except for Kaweah (n = 4). Most individuals were collected as adults, except for three populations where larvae were obtained from developing fruit (Boulevard, Grand Canyon, Santa Barbara). Adults were collected from one to several plants. Santa Barbara larvae were obtained from a single fruit and Grand Canyon and Boulevard larvae were collected from several fruits on two plants. Total genomic DNA was isolated from 30 adults and 16 larvae following a modified Harrison et al. (1987) protocol. Before extraction, the head, wings and genitalia were removed from adult specimens and kept as vouchers.
We evaluated mtDNA sequence variation by amplifying an 835-bp fragment within the COI subunit. We amplified the COI fragment for each individual in 30 µL reaction volumes using polymerase chain reaction (PCR; 1 × Promega PCR buffer, 2.5 mm MgCl2, 0.2 mm dNTPs, 0.25 µm each primer, 1 unit Taq polymerase and 10 ng of DNA template) for 35 cycles (60 s at 95 °C, 60 s at 52 °C, 90 s at 72 °C). Cycle sequencing products for forward and reverse strands were amplified from initial PCR products (50 ng of product, 0.4 µm primer and 4 µL of Thermo Sequenase Dye Terminator Sequencing Mix; Amersham Life Sciences) for 25 cycles (30 s at 96 °C, 30 s at 50 °C, 4 min at 60 °C) and sequenced on an ABI Prism 377 DNA sequencer. Of the 835-bp fragment amplified, we obtained 755-bp sequences that were readily aligned by eye as there were no indels. Representative sequences were deposited into GenBank (AF182761–AF182778).
Phylogeographic analyses were conducted using parsimony, likelihood and distance algorithms in paup* 4.0b1 (Swofford 2000). T. baccatella, T. treculeanella and the four species of the sister genus Parategeticula were chosen as outgroups based on prior phylogenetic analyses (Pellmyr & Leebens-Mack 1999). To simplify analysis, individuals bearing identical sequence haplotypes were condensed to a single haplotype designation.
We estimated the phylogeny with maximum parsimony using both simple and random addition in a heuristic search. Tree–bisection–reconnection (TBR) branch swapping and Multrees options were employed for both the likelihood and parsimony analyses. The maximum likelihood tree was generated with a heuristic search using the HKY85 model (Hasegawa et al. 1985) with estimated transition/transversion ratios. The shape parameter of the gamma distribution was set to α = 0.2 following estimates for other species of Tegeticula (Pellmyr & Leebens-Mack 1999). We also used neighbour joining to generate distance trees with several models of evolution: uncorrected p, Kimura 2-parameter and Jukes-Cantor with the shape parameter of the gamma distribution set to α = 0.2. Five hundred bootstrap replicates were generated to estimate support for the parsimony trees and 100 replicates were generated for the likelihood trees.
Results
We found 18 mtDNA haplotypes, differing at one to 15 nucleotide sites (0.13–1.99% sequence divergence). There were no indels inferred among the haplotypes and of the 755 sites examined, 32 were variable. The number of haplotypes per population ranged between one and five. Six of the eight study populations had multiple mtDNA haplotypes, whereas two (San Diego and Boulevard) were fixed for single haplotypes (Table 1). Only one haplotype was shared among multiple populations (haplotype ‘f’ in San Diego, Boulevard and Pinyon Flat). Haplotype diversity was as high in populations sampled from larvae within developing fruit as in populations that were collected as adult moths (Wilcoxon rank sums χ2 = 0.2301, 1 d.f., P = 0.6314).
| Population | Sample size | Haplotypes (n) |
|---|---|---|
| T. m. maculata | ||
| Kaweah, Tulare Co. (N36°32′ W118°55′) | 4 | a(3), b(1) |
| Kern Canyon, Kern Co. (N35°27′ W118°44′) | 6 | h(2), i(3), j(1) |
| Santa Barbara, Santa Barbara Co. (N34°44′ W119°59′) | 6 | n(2), o(1), p(1), q(1), r(1) |
| T. m. extranea | ||
| Boulevard, San Diego Co. (N32°40′ W116°17′) | 6 | f(6) |
| Jaraguay Pass, Baja California (N29°23′ W114°23′) | 6 | g(4), e(2) |
| Grand Canyon, Mohave Co. (N35°33′ W113°20′) | 6 | k(4), l(1), m(1) |
| Pinyon Flat, Riverside Co. (N32°34′ W116°28′) | 6 | c(1), d(1), f(4) |
| San Diego, San Diego Co. (N32°53′ W117°05′) | 6 | f(6) |
The parsimony analysis resulted in six most-parsimonious trees (36 steps, consistency index = 0.74, retention index = 0.86). The strict consensus tree indicated three distinct lineages: a Tegeticula maculata maculata clade (henceforth ‘Maculata’), a T. m. extranea (‘Extranea’) clade and another clade representing the two mtDNA haplotypes from the Kaweah population (‘Kaweah’). The bootstrap analysis provided 100% support for the major division among the three clades in the parsimony analysis. The likelihood and distance analyses produced the same tree topology. Both of these algorithms produced a tree with three clades identical to those of the parsimony analysis, with the exception that Kaweah was basal to the remainder of T. maculata (Fig. 3).
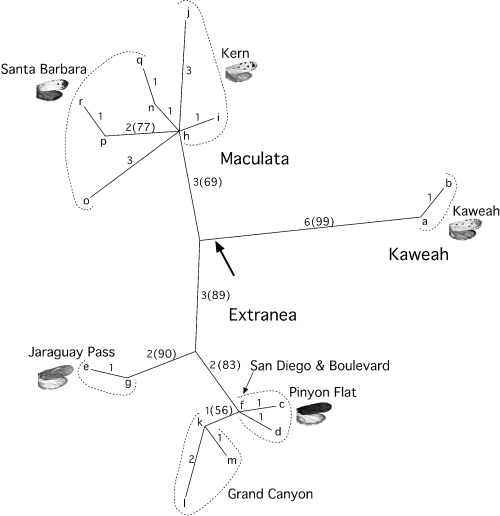
The single maximum likelihood tree (−ln likelihood = 1995.20) derived from cytochrome oxidase I (COI) mitochondrial DNA (mtDNA) sequences of Tegeticula maculata. The letters represent mtDNA haplotypes. The numbers near each branch indicate the number of nucleotide substitutions along that branch, the numbers in parentheses are bootstrap values and the arrow indicates the position of the root.
Because the split between the Maculata and Kaweah clades was unexpected, we tested the strength of this division by constructing a constraint tree forcing Kaweah and Maculata haplotypes into a single clade. The resulting trees were not significantly different from the original tree topology (Kishino-Hasegawa test P = 0.96). Moreover, trees where Kaweah and Extranea were forced as a monophyletic group were also not significantly different from the original topology (Kishino-Hasegawa test P = 0.86). The placement of Kaweah was uncertain, but all analyses distinguished these haplotypes from Maculata and Extranea.
Sequence divergence was similar among the three clades. Haplotypes from Kaweah differed from the Extranea haplotypes by a mean of 1.67 ± 0.036% (standard error) sequence divergence and from the Maculata haplotypes by a mean of 1.46 ± 0.034%. The Maculata and Extranea haplotypes differed from each other by a mean sequence divergence of 1.37 ± 0.021%.
To provide an approximate estimate of the divergence time for the three lineages, we estimated the likelihood tree assuming a molecular clock. We did not reject the clock model (likelihood ratio test: χ2 = 28.924, d.f. = 22, P > 0.05). Using the node to tip heights provided in paup*, divergence times were estimated for the split between Kaweah and Extranea/Maculata and for the split between Extranea and Maculata. The relative age was defined as the height of the node of interest divided by the height of the split between T. maculata and other Tegeticula. The relative age was multiplied by 41.7 Myr, the approximate age of the root as determined by Pellmyr & Leebens-Mack (1999). We estimated that the Kaweah mitochondrial lineage diverged from the remainder of T. maculata about 9.9 Ma and Maculata and Extranea split about 9.1 Ma. Using the same methodology, we also estimated the divergence time of the oldest split within Extranea; the divergence of Jaraguay Pass and other Extranea haplotypes occurred approximately 4.7 Ma. These divergence times are subject to error and should be taken with caution (Bromham et al. 2000). We have included these estimates here only with the intention of approximating whether we are considering shorter (thousands of years) or longer (millions of years) timescales.
Even within clades, there was considerable substructure (Fig. 3). Jaraguay Pass and Grand Canyon were distinguished as monophyletic groups within Extranea, suggesting restricted gene flow between these and more geographically distant populations. Maculata also exhibited substructure; despite high overall haplotype diversity, there were no shared haplotypes in our samples.
Discussion
Phylogeographic structure within Tegeticula maculata
As predicted, T. m. maculata and T. m. extranea are highly diverged lineages that split approximately 9–10 Ma. This deep split was shown earlier by the sequence divergence between single moths of each subspecies that were included in a phylogeny of the Prodoxidae (Pellmyr & Leebens-Mack 1999). Surprisingly, we detected a second split within T. m. maculata that was as old as the division between the subspecies. This divergence between Kaweah and Maculata was unexpected, as there are no described morphological differences between them. Wing maculation is variable among T. m. maculata moths and was used previously to delineate a third subspecies, apicella (Powell & Mackie 1966). Subspecies apicella, however, does not strictly correspond with either of the lineages within T. m. maculata and would be a polyphyletic group. Both the Kaweah and Kern populations were labelled apicella (Powell & Mackie 1966), but Kern haplotypes cluster with haplotypes from the Santa Barbara population. The factors restricting gene flow between Kaweah and nearby Maculata populations are unknown, but the population of Hesperoyucca whipplei at Kaweah may currently be isolated from its nearest neighbours by as much as 50 km (Haines 1941; Powell & Mackie 1966). For Extranea populations, there were shared haplotypes between populations further apart than 50 km, but there is reason to believe that interspersed host plants may act as a bridge between them (Powell & Mackie 1966; CalFlora Database 2000).
Another possible explanation for the division between Kaweah and Maculata is lineage sorting. Sorting could explain why the current distribution of mtDNA haplotypes reflects an old split between two morphologically indistinguishable lineages. This type of process was proposed in a phylogeographic assessment of extant and permafrost-preserved brown bears (Leonard et al. 2000). Extant bears in North American populations represent geographically structured haplotypes from three clades, suggesting there were three independent invasions from the Old World. However, the distribution of mtDNA haplotypes of permafrost-preserved specimens indicates that there was probably a single invasion and that the current phylogenetic structure was a result of a subsequent loss of haplotypes since the Pleistocene. The current phylogeographic structure, then, may present a misleading view of the processes generating that structure (Leonard et al. 2000).
As with T. m. maculata, a similar pattern of disjunct genetic structure is also found in Greya politella, another prodoxid moth, associated with several plant species in the Saxifragaceae (Brown et al. 1997). G. politella collected from the same site at Kaweah were highly differentiated from other G. politella populations in California and the Kaweah population was as genetically distinct from other California populations as the major clades separating northern (Washington and Idaho) and southern (California) populations. The factors generating parallel genetic structure in these two species are unknown, but the striking similarity suggests similar patterns of historical biogeography and holds interesting prospects for further investigations in this region.
Phylogeographic structure within clades
The overall geographic structure of populations within T. maculata suggests isolation and potential for further divergence within each of the three clades, especially for Extranea. For instance, the Grand Canyon population is presently isolated by at least 270 km from its nearest neighbouring population. Evidence from subfossil H. whipplei leaves in packrat middens indicates that the Grand Canyon population is a relict of a more continuous distribution of H. whipplei throughout the Sonoran and Mojave deserts (Fig. 2). H. whipplei disappeared from the Death Valley and the lower Colorado River Valley regions at the end-glacial period 11 000–12 000 years bp (King & Van Devender 1977; Cole 1986; Woodcock 1986; Van Devender 1990), isolating the Grand Canyon population as summer temperatures elsewhere increased beyond the plant’s tolerance range (Woodcock 1986). Hence, Grand Canyon moths have probably been isolated from the remainder of Extranea for at least 11 000 years as a result of this climate change (Wells & Woodcock 1985). Grand Canyon’s phylogenetic affinity to southern California moths may be explained by the recent biogeographic connection with cismontane T. m. extranea via now-extinct Colorado River Valley host populations.
The morphologically distinct Jaraguay Pass population in central Baja California is also highly diverged from the rest of Extranea. Jaraguay Pass split from Extranea early in its history, perhaps as long as 4.7 Ma. The Jaraguay Pass population is in an isolated segment of the Sonoran desert on the Baja California peninsula, separated from more northern populations by about 80 km. This discontinuity between northern and southern populations corresponds with the boundary of the Sonoran desert (Fig. 2) and the intervening region is characterized as a chaparral–desert transition zone with different climatic regimes (Wiggins 1980). If the southern Baja California populations are relics of the plant’s past extensive Sonoran desert distribution, the other surviving population at Pinacate in northwestern Sonora (Turner et al. 1995) should contain the closest relatives of Jaraguay Pass.
Conclusions
The substantial genetic divergence between the morphologically defined subspecies leads to the conclusion that they are potentially separate species, especially as the sequence divergence between them is nearly as large as it is among the majority of species in the Tegeticula yuccasella complex (Pellmyr & Leebens-Mack 2000). We refrain from formally delineating species, however, because of the equally deep split within T. m. maculata, which is not accompanied by corresponding morphological divergence and also because we lack evidence for genetic differentiation in areas of sympatry. Further genetic sampling and ecological work is required for a robust answer to the number of species within the current T. maculata.
The high haplotype diversity and phylogeographic structure among populations within subspecies is indicative of considerable isolation among patchily distributed habitats and would seem to be a good candidate for allopatric diversification (Mayr 1963; Coyne 1992). It is interesting then that the T. maculata lineage which has existed since a point in time near the origin of the plant–pollinator mutualism at least 35–40 Ma, only appears to have given rise to three clades that have persisted into the present. The climatic history of the arid–semiarid southwestern North America, with repeated contractions and expansions of biogeographic regions, may have played a role in reducing the probability that locally diversifying lines speciate. The subfossil record shows that the changing Hesperoyucca whipplei distribution is tied to changing biogeographic boundaries during and after glacial periods. The patchy distribution observed today is a remnant of a past, more contiguous range (Fig. 2) and the high structure among moth populations may, to a fair extent, have resulted from lineage sorting of ancient mtDNA haplotypes followed by local mitochondrial differentiation. This scenario can explain both high levels of sequence divergence and the scarcity of adaptive diversification over longer time spans.
Acknowledgements
We thank Manuel Balcázar-Lara, Virginia Boucher, Nancy Brian, John Brown, Neil Cobb, Adolfo Ibarra, Jim Leebens-Mack, Clay Nelson, Jerry Powell, Kelly Richers, Joe Shannon, Larry Stevens and John Thompson for collecting moths and providing site information. We are indebted to Jerry Powell for information regarding the Baja California moths. Grand Canyon National Park provided permission to gather moth samples. David Althoff and Jim Leebens-Mack provided helpful advice on phylogenetic analysis. We thank David Althoff, Beau Crabb, Daniel Funk, Deborah Marr and Michel Solignac for making helpful comments on this manuscript. This project was funded by NSF grants DEB 9528072 and DEB 9509056 and a grant from the National Geographic Society.
References
The research presented is part of a long-term project examining the phylogenetic and phylogeographic relationships of yucca moths and their host plants to understand the origins of this interaction and to understand further how specialization and co-evolution shape patterns of diversification on different timescales. Kari Segraves is a graduate student studying the evolution and ecology of plant–pollinator interactions. Olle Pellmyr integrates molecular phylogenetics and ecological tools to address questions about species interactions, the evolution of mutualisms and co-evolution.




