The use of ‘touchdown’ polymerase chain reaction increases the sensitivity and specificity of t(11;14)(q13;q32) detection in patients with mantle cell lymphoma
Mantle cell lymphoma (MCL) is a distinct form of non-Hodgkin's lymphoma characterized by the t(11;14)(q13;q32) translocation which results in the over-expression of the cyclin D1 (CCND1, PRAD1, BCL-1) gene (Swerdlow et al, 2001). The presence of the t(11;14)(q13;q32) translocation may be determined by a variety of techniques with varying degrees of success. There are several polymerase chain reaction (PCR) procedures already available (Williams et al, 1993; Rimokh et al, 1994; Fan et al, 1998). However, specificity can be a problem with these types of PCR methods (Rimokh et al, 1994). Touchdown PCR (Td-PCR; Don et al, 1991) was developed to help eliminate the production of spurious PCR products. We report the use of Td-PCR for detection of the t(11;14)(q13;q32).
Genomic DNA samples from four patients with MCL and one healthy volunteer (all individuals gave informed consent) were amplified using both conventional [hold (95°C 15 min) 30 cycles (94°C 60 s; 50°C 60 s; 72°C 50 s) and hold (72°C 7 min; 4°C∞)] and touchdown [hold (95°C 15 min) 15 cycles (94°C 60 s; 65°C 60 s; 72°C 50 s; reducing the annealing temperature by 1°C each cycle) 20 cycles (94°C 60 s; 50°C 60 s; 72°C 50 s) and hold (72°C 7 min; 4°C∞)] thermal cycler programming (1°C/s ramp rate). The PCR mix (25 µl) included: Thermo-Start DNA polymerase (0·5 U; ABgene, UK) and buffer (1 x; without magnesium chloride); magnesium chloride (1·5 mmol/l; ABgene) dNTP mix (0·2 mmol/l; ABgene) primers (10 µmol/l each; Invitrogen, UK) MTC-A (5′-CCT CTC TCC AAA TTC CTG-3′; Williams et al, 1993) and LJH (5-TGA GGA GAC GGT GAC C-3′; Brisco et al, 1990); template DNA (2 µl) and nuclease-free water (Promega UK, UK). A GeneAmp 9700 thermal cycler (Applied Biosystems, UK) was used. All PCR products were analysed by Tris-borate EDTA (TBE) agarose (2% w/v) gel electrophoresis. The identity of each Td-PCR product was confirmed by cycle sequencing.
The two techniques produced different levels of PCR specificity with the same DNA samples (Fig 1). Where conventional programming was used, many of the DNA samples produced a spurious band [approximately 100 base pairs (bp) in size] that was absent when touchdown programming was used. The majority of MCL DNA samples, regardless of the thermal cycler programming used, produced a band (approximately 600 bp in size). Sequencing of the touchdown PCR products revealed these bands shared a high degree homology with available t(11;14) breakpoint sequences (data not shown). Sample MCL-S5 yielded a high-molecular-weight band only under touchdown conditions. The normal control sample produced detectable bands only under conventional conditions. The water control produced no bands.
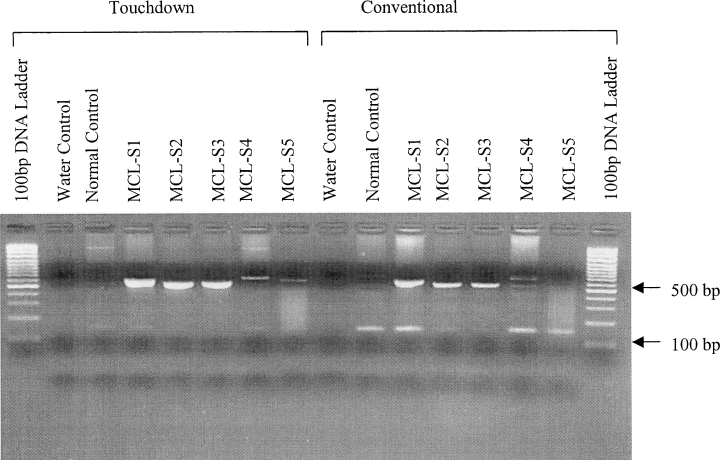
The ethidium-bromide-stained agarose (2% w/v) electrophoresis gel of the PCR products generated by touchdown and conventional thermal cycler programming. The sample (10 µl) was mixed with loading dye (1 µl) before loading. The DNA samples MCL-S5 and MCL-S4 were isolated from bone marrow, MCL-S1 from blood, and MCL-S2 and MCL-S3 from frozen tissue samples. The normal control DNA sample was isolated from blood. The 100 bp DNA ladder (1 µl; GeneRuler 100 bp DNA Ladder Plus; MBI Fermentas, Lithuania) was used according to the manufacturer's recommendations.
We have demonstrated that it is possible to increase the level of specificity by changing the method used to program a thermal cycler, as evidenced by sample 5. A possible reason for this observed increase in sensitivity could be the lack of competition between the real and spurious PCR products for the available primers and enzymes (as seen with conventional thermal cycler programming), thus allowing more specific amplification and, in turn, increased product yield. Therefore, the apparent increase in sensitivity was achieved by increasing the specificity. The Td-PCR procedure is capable of detecting t(11;14) breakpoints in genomic DNA samples diluted to 25 pg or 10 cell equivalents (data not shown), which implies that this method is no more sensitive than other PCR systems. In summary, the application of an alternative thermal cycling program for the detection of the t(11;14) can increase the specificity (and to some degree the sensitivity) of this PCR method.




