Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera
Abstract
Phylogenetic relationships among members of the Mecoptera and Siphonaptera were inferred from DNA sequence data. Four loci (18S and 28S ribosomal DNA, cytochrome oxidase II and elongation factor-1α) were sequenced for 69 taxa selected to represent major flea and mecopteran lineages. Phylogenetic analyses of these data support a paraphyletic Mecoptera with two major lineages: Nannochoristidae + (Siphonaptera + Boreidae) and Meropidae + ((Choristidae + Apteropanorpidae) (Panorpidae + (Panorpidae + Bittacidae))). The flea family Ctenophthalmidae is paraphyletic, and the Ceratophylloidea is monophyletic. Morphological evidence is discussed which is congruent with the placement of Siphonaptera as sister group to Boreidae.
Introduction
Mecoptera is a small holometabolous insect order with approximately 600 extant described species placed in nine families and 32 genera (Penny & Byers 1979; Penny 1997). This group is called scorpionflies because the male ninth abdominal (genital) segment of one family (Panorpidae) is enlarged, bulbous, and curves anterodorsally, resembling the stinger of a scorpion. Two families — Panorpidae and Bittacidae — contain 90% of mecopteran species. Panorpidae (377 spp.) is the most speciose family with three described genera: Panorpa (254 spp.) is distributed throughout northern continents and Indonesia, but not in Australia; Neopanorpa (110 spp.) is distributed throughout India, southern China, Indochina and southward to Java and Borneo; and Leptopanorpa (13 spp.) is restricted entirely to Java (Byers & Thornhill 1983). Bittacidae, sometimes known as hangingflies because species hang from plants by the fore or mid legs, comprises 172 species placed in 16 genera. During courtship, males present females with a nuptial meal, and in some species males mimic females to steal the nuptial meal (Thornhill 1979). Bittacidae is the most diverse neotropical mecopteran group, where the ranges of the small genera, Anabittacus (1 sp.), Issikiella (5 spp.), Kalobittacus (8 spp.), Nannobittacus (4 spp.), Neobittacus (2 spp.) and Pazius (8 spp.), overlap within the ranges of neotropical Bittacus (25 spp.). Orobittacus, Apterobittacus and Hylobittacus are monotypic genera restricted to North America, and there are seven additional Bittacus species in North America. Ten species of Harpobittacus and one each of Austrobittacus, Edriobittacus, Symbittacus and Tytthobittacus are endemic to Australia. Anomalobittacus (1 sp.) and 48 species of Bittacus are restricted to Africa, and comprise the entire mecopteran fauna of Africa (Byers 1991). The remaining Bittacus species occur in Europe, Japan, Korea, India, Taiwan, China and Thailand (Penny 1997).
The other mecopteran families, although less speciose, show a spectacular degree of variation in morphology and ecology. Boreidae (snow fleas) is a small group of 26 species placed in three genera that is distributed throughout North America, Europe and Japan. Adults emerge in winter and are associated with bryophytes (Penny 1977; Russell 1982). Wings are reduced to small, oval flaps in females, and thin spiny hooks in males, which function to clasp the female during mating. Boreids are unique among Mecoptera in their ability to jump up to 30 cm when disturbed, which not only facilitates escape from predators, but also allows them to cross light, fluffy snow where it is difficult to walk (Penny 1977). In the case of Hesperoboreus, the male jumps directly onto the female prior to copulation (Cooper 1972). Panorpodidae, which morphologically resembles Panorpidae except for a much shorter rostrum, consists of two genera, Brachypanorpa in the Pacific north-western USA (3 spp.) and in Appalachia (2 spp.), and Panorpodes (4 spp.) occurring in Japan. Choristidae consists of 10 species in three genera restricted entirely to Australia, while Nannochoristidae comprises two genera and seven species found in Australia and South America. Meropeidae, ‘earwig flies’, consists of two extant species: Merope tuber (eastern North America) and Austromerope poultoni (Australia), both of which are cockroach-like in general appearance with extremely large forcep-like appendages on the abdomen. Eomeropidae is also cockroach-like and is a monotypic family with one Chilean species, Notiothauma reedi. Apteropanorpidae, another apterous mecopteran family adapted to cold climates, has two species known from Tasmania (Byers & Yeates 1999).
The monophyly of each mecopteran family is well established by morphological characters that have been summarized in other studies (Kaltenbach 1978; Willmann 1987; Byers 1991). From a morphological standpoint, some of the families appear to be living fossils (e.g. Eomeropidae and Meropeidae) and may be the sole remnants of what were once more diverse lineages (Kaltenbach 1978; Willmann 1989). Mecoptera have a very well-documented fossil history and are among the most conspicuous part of the insect fauna of the Lower Permian. There are 348 species of Mecoptera described from the Permian, Mesozoic and Tertiary, representing 87 genera in 34 families (see Willmann 1977, 1981 1983, 1984a,b, 1987). There is no other holometabolous insect order that has such a biased distribution of species within families, where 90% of the species occur in ~20% of the families, or where the diversity of the extinct taxa at the familial and generic level is about three times that of the extant taxa.
Siphonaptera (fleas) is a highly specialized holometabolous insect order with 2380 described species placed in 15 families and 238 genera (Lewis & Lewis 1985). Fleas are laterally compressed, wingless insects that range from 1 to 10 mm in length. The head is usually small and shield- or helmet-shaped, compound eyes are absent, and mouthparts are specialized for piercing and sucking (Dunnet & Mardon 1991). Fleas are entirely ectoparasitic, with ~100 species as parasites of birds and the remaining species as parasites of mammals (Holland 1964). Flea distribution extends to all continents, including Antarctica, and fleas inhabit a range of habitats and hosts from equatorial deserts, through tropical rainforests, to the arctic tundra. Fleas are of tremendous economic importance as vectors of several diseases important to human health, including bubonic plague, murine typhus and tularaemia (Dunnet & Mardon 1991).
From a phylogenetic standpoint, Siphonaptera is the most neglected of the holometabolous insect orders. While we have a reasonable knowledge of flea taxonomy at the species and subspecific level, and a relatively good record of their biology and role in disease transmission, phylogenetic relationships among fleas at any level have remained virtually unexplored. Classically, the major obstacle in flea phylogenetics has been their extreme morphological specializations associated with ectoparasitism, and the inability of systematists to adequately homologize characters across taxa. The majority of characters used for species diagnoses are based on the shape and structure of their extraordinarily complex genitalia, or the presence and distribution of setae and spines (Traub & Starcke 1980; Dunnet & Mardon 1991). While these characters are adequate for species diagnoses, they are mostly autapomorphic at the species level and of limited utility for phylogenetic reconstruction. Siphonaptera appears to have many instances of parallel reductions and modifications, probably associated with multiple invasions of similar hosts, which may obscure homology (Holland 1964).
Ordinal phylogeny
While it is clear that Mecoptera and Siphonaptera are holometabolous insect orders, their position relative to the other Holometabola is somewhat controversial. Hennig (1969) placed Mecoptera as sister group to Diptera in Antliophora, but was uncertain as to whether Siphonaptera should be included within Antliophora, or even affiliated with the other mecopteroid orders. Based on similarities of the proventriculus, Ross (1965) argued for a sister group relationship between Mecoptera and Siphonaptera. Alternatively, Boudreaux (1979) placed Mecoptera as sister group to Diptera + Siphonaptera. Kristensen (1981, 1991) favoured a sister group relationship between Mecoptera and Siphonaptera. The sister group to Antliophora is probably Amphiesmenoptera (Lepidoptera + Trichoptera) (Whiting et al. 1997; Kristensen 1999). The close association between Mecoptera and Siphonaptera has been borne out in recent molecular studies (Chalwatzis et al. 1996; Whiting et al. 1997; Whiting 2001, 2002), although the monophyly of Antliophora + Amphiesmenoptera is not well supported by DNA sequence data (see Whiting 2002).
Familial phylogeny
The phylogeny of Mecoptera has centred around two problematic families: Nannochoristidae and Boreidae. The Nannochoristidae have unusual, aquatic larvae (Pilgrim 1972), a pigmented larval ‘eye spot’ (Melzer et al. 1994), unique venational characteristics (Kristensen 1989) and a suite of characters that are presumably primitive for Mecoptera (Willmann 1987). Phylogenetically, Nannochoristidae was placed as the most basal mecopteran family (Willmann 1987), sister group to Diptera + Siphonaptera (Wood & Borkent 1989) and even elevated to ordinal status, ‘Nannomecoptera’ (Hinton 1981). The Boreidae also have unusual morphological features (Penny 1977) and were placed as a highly derived mecopteran sister group to Panorpodidae (Penny 1975), as a relatively basal group placed in a trichotomy with Meropeidae and Panorpomorpha (Willmann 1987: Fig. 1) or elevated to their own order, ‘Neomecoptera’ (Hinton 1958). Hinton’s suggestion that Nannochoristidae and Boreidae should be given their own ordinal status was based exclusively on a phenetic argument, essentially that these taxa appear so different from other Mecoptera that they deserve ordinal status.
Penny (1975) presented an ‘intuitive’ phylogeny in which Meropeidae is the basal-most taxon with Boreidae placed as sister group to Panorpodidae. Mickoleit (1978) inferred familial relationships based on characters of genitalia, and proposed a phylogeny in which the Nannochoristidae and Bittacidae are the basal-most taxa (Fig. 1a). Kaltenbach (1978) presented Mecoptera subdivided into three suborders, Protomecoptera (Meropeidae + Eomeropidae), Neomecoptera (Boreidae) and Eumecoptera (remaining families), but did not present a specific phylogeny for these taxa. In a comprehensive analysis of mecopteran morphology from extinct and extant taxa, Willmann (1987, 1989) presented a phylogeny in which Nannochoristidae is the basal-most taxon, with Panorpidae + Panorpodidae forming the most apical clade (Fig. 1b). This phylogeny was not the result of a formal quantitative analysis of a coded character matrix, but Willmann did provide an explicit explanation of the characters supporting each node of the phylogeny. In all cases, these authors are uncertain as to the placement of Meropeidae, and it is possible that its close association with Eomeropidae (i.e. Protomecoptera sensu Kaltenbach) is due to symplesiomorphy.
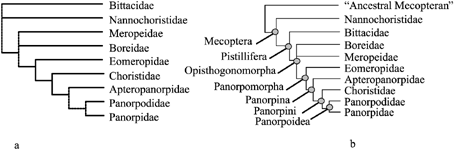
Phylogeny of Mecoptera based on morphology after Mickoleit (1978) (a) and Willmann (1989) (b).
Familial relationships among fleas are much less well resolved and have been less studied than mecopteran families. There is no generally accepted higher classification for Siphonaptera, and several classifications published in recent years have significantly conflicting treatments of superfamilial relationships (Mardon 1978; Smit 1979, 1983, 1987; Traub & Starcke 1980; Traub et al. 1983; Lewis & Lewis 1985; Dunnet & Mardon 1991). The monophyly of many flea families is questionable, and certain families that have been used as a catch-all for a wide range of divergent taxa (e.g. Ctenophthalmidae) are almost certainly paraphyletic assemblages. The phylogeny presented by Smit (1979: Fig. 2) is not based on a formal quantitative analysis of flea morphology, and the monophyly of each of these groups is questionable.
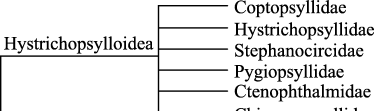
Phylogeny of Siphonaptera after Smit (1979).
Materials and methods
Sequence data were generated for a total of 69 taxa, representing Amphiesmenoptera (six taxa), Diptera (three taxa), Mecoptera (41 taxa) and Siphonaptera (19 taxa). Although there is morphological and molecular evidence to support the placement of Strepsiptera within Antliophora (Whiting 1998), Strepsiptera was excluded as an outgroup in this analysis because of the difficulty of accurately sequencing the protein-coding genes for strepsipteran exemplars. All mecopteran families, with the exception of Eomeropidae, and the majority of flea families (nine of 15) are included in this analysis (Appendix 1). Thoracic muscle tissue was dissected and incubated in a standard buffer (100 mm ethylenediaminetetraacetic acid (EDTA), 10 mm Tris, 1% sodium dodecylsulphate (SDS), 20 µg proteinase K, pH 7.5) overnight at 55 °C. After buffer incubation, DNA was extracted using standard phenol/chloroform extraction protocols and concentrated by column purification (Centricon-30, Ambion). Four genes were targeted for amplification and sequencing: 18S ribosomal DNA (18S rDNA), 28S ribosomal DNA (28S rDNA), elongation factor-1α (EF-1α) and cytochrome oxidase II (COII). Primer sequences are given in Table 1; relative primer positions and cycling conditions are given in Fig. 3. Genomic DNA templates and controls were amplified using standard polymerase chain reaction (PCR) techniques in a Perkin-Elmer 9600 thermocycler. Product yield, specificity and potential contamination were monitored by agarose gel electrophoresis. The target product was purified and cycle-sequenced using the ABI dRhodamine cycle sequencing kit. The sequencing reactions were column purified and analysed with the ABI 377 automated sequencer. In all cases, DNA was sequenced from complementary strands, with sufficient overlap for the larger genes to ensure the accuracy of all sequence output. Manual correction of chromatography data was facilitated by the program Sequencher™ 3.1.1 (Genecodes 1999), which automatically aligns chromatographs of the sequence output to provide more efficient and accurate sequence correction.
| Primer | Sequence (5′ → 3′) |
|---|---|
| 18S 1.2F | TGCTTGTCTCAAAGATTAAGC |
| 18S ai | CCTGAGAAACGGCTACCACATC |
| 18S a0.7 | ATTAAAGTTGTTGCGGTT |
| 18S a0.79 | TTAGAGTGCTYAAAGC |
| 18S a1.0 | GGTGAAATTCTTGGAYCGTC |
| 18S a2.0 | ATGGTTGCAAAGCTGAAAC |
| 18S a3.5 | TGGTGCATGGCCGYTCTTAGT |
| 18S 7F | GCAATAACAGGTCTGTGATGCCC |
| 18S 9R | GATCCTTCCGCAGGTTCACCTAC |
| 18S 7R | GCATCACAGACCTGTTATTGC |
| 18S bi | GAGTCTCGTTCGTTATCGGA |
| 18S b0.5 | GTTTCAGCTTTGCAACCAT |
| 18S b2.5 | TCTTTGGCAAATGCTTTCGC |
| 18S b3.0 | GACGGTCCAACAATTTCACC |
| 18S b3.9 | TGCTTTRAGCACTCTAA |
| 18S b5.0 | TAACCGCAACAACTTTAAT |
| 18S b7.0 | ATTTRCGYGCCTGCTGCCTTCCT |
| 28S rD1.2a | CCCSSGTAATTTAAGCATATTA |
| 28S rD3.2a | AGTACGTGAAACCGTTCASGGGT |
| 28S A | GACCCGTCTTGAAGCACG |
| 28S Rd4.2a | CTAGCATGTGYGCRAGTCATTGG |
| 28S Rd4.5a | AAGTTTCCCTCAGGATAGCTG |
| 28S Rd4.8a | ACCTATTCTCAAACTTTAAATGG |
| 28S rD5a | GGYGTTGGTTGCTTAAGACAG |
| 28S Rd6.2a | GAAAGGGAATCYGGTTMMTATTCC |
| 28S rD7b1 | GACTTCCCTTACCTACAT |
| 28S Rd6.2b | AATAKKAACCRGATTCCCTTTCGC |
| 28S rD5b | CCACAGCGCCAGTTCTGCTTAC |
| 28S B | TCGGAAGGAACCAGCTAC |
| 28S Rd4.2b | CCTTGGTCCGTGTTTCAAGACGG |
| 28S Rd3.2b | TGAACGGTTTCACGTACTMTTGA |
| COII-2a | ATAGAKCWTCYCCHTTAATAGAACA |
| COII-9b | GTACTTGCTTTCAGTCATCTWATG |
| COII-F-leu | TCTAATATGGCAGATTAGTGC |
| COII-R-lys | GAGACCAGTACTTGCTTTCAGTCATC |
| EF-1α M 44–1 | GCTGAGCGYGARCGTGGTATCAC |
| EF-1α M 46–1 | GAGGAAATYAARAAGGAAG |
| EF-1α M 52.7 | GTCAAGGARYTGCGTCGTGG |
| EF-1α rcM 4.0 | ACAGVCACKGTYTGYCTCATRTC |
| EF-1α rcM 53.2 | GCAATGTGRGCIGTGTGGCA |
| EF-1α rcM 53.0 | ATRTGRGCNGTGTGGCAATC |
| EF-1α rcM 52.6 | GCYTCGTGGTGCATYTCSAC |
| EF-1α rcM 51–1 | CATRTTGTCKCCGTGCCAKCC |
| EF-1α rcM 44.9 | CTTGATGAAATCYCTGTGTCC |
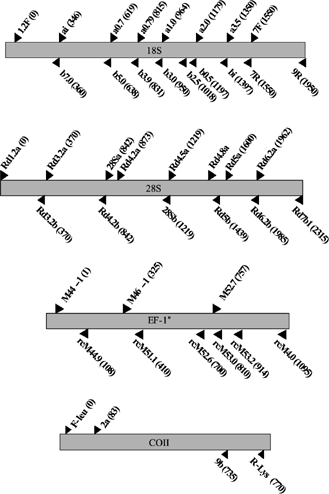
Map of primer positions for 18S rDNA, 28S rDNA, EF-1α and COII used in this study. Primer sequences are given in Table 1.
Sequences were assembled in Sequencher™ 3.1.1 (Genecodes 1999). The protein-coding genes (COII and EF-1α) were manually aligned with reference to the amino acid sequences. For the ribosomal genes, a gross alignment was performed by manually aligning the conserved domains across the taxa. Conserved domains, and variable regions between domains, were removed in sections and entered into the computer program poy (Gladstein & Wheeler 1999) to undergo more exhaustive alignment. poy was implemented on a dedicated parallel cluster (64 CPUs, 500 mHz with 1 GB RAM) using gap cost = 2, change cost = 1, with TBR (Tree Bisection and Reconnection), branch swapping on 100 alignments, with the option ‘implied alignment’ implemented. While poy is designed to construct a topology while simultaneously performing alignment (Wheeler 1999), the implied alignment option yields a multiple alignment which is more optimal than those typically found by other alignment algorithms, such as malign (Wheeler & Gladstein 1994) or Clustal W (Thompson et al. 1994). Variable alignment regions which appeared ambiguously aligned between the ingroup and outgroups, but relatively conserved within each family, were aligned independently within each mecopteran family using poy with the parameters as described above. These variable regions were excluded from the outgroups because resolution among these taxa is not the focus of this study. Each of these regions was considered an alignment block, and the blocks were assembled into a single matrix by scoring the taxa outside the block with missing values, as described elsewhere (see Whiting 2001, 2002). The alignment can be found at http://dnasc.byu.edu/~whitinglab.
Trees were reconstructed under parsimony with gaps treated as missing data using the program nona (Goloboff 1994) with 50 random addition sequences and TBR branch swapping. Partitioned Bremer support values (Baker & DeSalle 1997) were calculated using the program TreeRot (Sorenson 1999) and paup*4.0 (Swofford 2000). The incongruence length difference (ILD) test was performed using the program arn with 1000 replications, and uninformative characters were removed (Farris et al. 1994). Trees were reconstructed with the variable blocked regions included and excluded from the analysis and under a variety of codon weighting schemes (1 : 1 : 0, 1 : 1 : 1, 3 : 5 : 1, and estimated values 5 : 10 : 1 (COII) and 2 : 4 : 1 (EF-1α)) to explore the sensitivity of the phylogenetic results to different weighting parameter values.
Results and discussion
Alignment of the sequence data for 18S resulted in 2137 characters, 522 of which were parsimony informative with one variable blocked region. Hypervariable regions of the alignment (positions 1545–1591 and 1617–1699) were excluded from the analysis. The 28S data consisted of a 6464 base pair (bp) alignment with eight variable blocked regions. The more conserved regions totalled 2114 bp, 739 of which were parsimony informative. The variable blocked regions consisted of 4350 bp, 450 of which were parsimony informative. Hypervariable regions of the alignment (positions 5741–5763, 7053–7084, 7215–7300 and 7527–8222) were excluded from the analysis. The EF-1α data consisted of 1092 bp, 415 of which were parsimony informative, with nucleotide 1 (nt1) = 58 (14%), nt2 = 30 (7%) and nt3 = 327 (78%). The COII data consisted of 599 bp, 326 of which were parsimony informative, with nt1 = 94 (29%), nt2 = 45 (14%) and nt3 = 187 (57%). Results of the ILD test failed to reject the hypothesis of data set incongruence for all combinations except for 18S vs. the protein-coding genes (Table 2). However, as the test was not symmetric (i.e. 18S and 28S were congruent, 28S and the protein-coding genes were congruent, but 18S and the protein-coding genes were incongruent), and because the ILD confounds incongruence due to conflicting signals with incongruence due to homoplasy (Dolphin et al. 2000), the molecular data sets were combined in a total evidence analysis.
| Partition comparison | α value |
|---|---|
| 28S/18S | 1.000 |
| EF-1α/28S | 1.000 |
| EF-1α/COII | 1.000 |
| EF-1α/18S | 0.001* |
| COII/18S | 0.001* |
| COII/28S | 0.194 |
| 18S/COII + EF-1α | 0.001* |
| 18S/COII + EF-1α + 28S | 1.000 |
| 18S + 28S/COII + EF-1α | 0.230 |
- * Values of α < 0.050 indicate sufficient evidence to reject the hypothesis of data set congruence.
Analysis of the 18S rDNA data, with variable blocked regions included, results in a topology where familial relationships are entirely unresolved, except for Panorpidae + Panorpodidae (Fig. 4). These data provide some resolution within the Panorpidae and Ceratophylloidea, but do not provide evidence for the paraphyly of any mecopteran family. Exclusion of the variable blocked regions results in a nearly identical topology. Analysis of the 28S rDNA data results in a topology where Meropeidae is the basal-most clade and Boreidae is sister group to Nannochoristidae + Siphonaptera (Fig. 4). Exclusion of the variable blocked regions results in a less resolved topology, but one which retains the clades (Boreidae (Nannochoristidae + Siphonaptera)) (Panorpidae (Bittacidae + Panorpodidae)), and a basal placement of Meropeidae. Analysis of the COII data for all nucleotide schemes investigated results in topologies which support Boreidae + Siphonaptera as the basal-most clade, with Nannochoristidae in a more derived position (Fig. 4). All COII analyses, rather surprisingly, also support a paraphyletic Panorpidae. Analysis of the EF-1α data with all nucleotide positions weighted equally supports a topology in which fleas, boreids and Meropeidae form a clade, although the first two groups are grossly paraphyletic in respect to each other (Fig. 4). Exclusion of third position nucleotides results in overall less resolution, although relationships among the fleas are fully resolved and more congruent with the other genes.
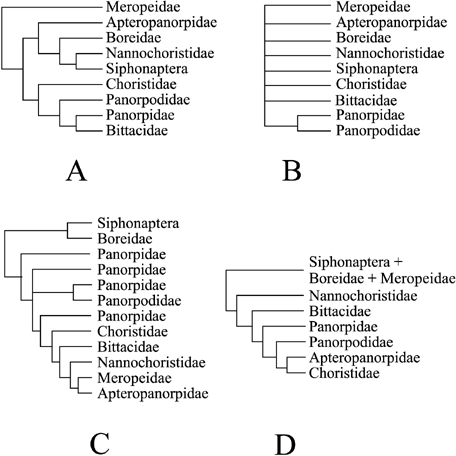
Summary trees for individual genes used in this analysis based on parsimony analysis: (A) 28S rDNA; (B) 18S rDNA; (C) COII; (D) EF-1α. The 18S tree is based on the entire alignment (conserved and variable regions) and is the strict consensus of 599 trees (L = 1506, CI = 0.58, RI = 0.82). The 28S tree is based on the entire alignment and is the strict consensus of 16 trees (L = 4276, CI = 0.52, RI = 0.81). The COII tree is based on equal weighting of all positions and is the strict consensus of nine trees (L = 2968, CI = 0.30, RI = 0.60). The EF-1α tree is based on equal weighting of all positions, generating only one tree (L = 3436, CI = 0.24, RI = 0.58).
Phylogenetic analysis of a single gene across the Mecoptera and Siphonaptera appears to be insufficient to resolve the phylogeny of these taxa. 18S results in a poorly resolved topology, COII results in a topology where Panorpidae is paraphyletic, EF-1α results in a topology where Boreidae and Siphonaptera are paraphyletic and 28S produces a topology where Meropeidae is the basal-most taxon and Apteropanorpidae is the sister group to fleas + boreids + nannochoristids. Indeed, the topologies from the individual genes are less congruent with phylogeny based on morphology than is the total evidence topology. Summing the Bremer and partitioned Bremer support values for various nodes on the topology reveals at what level the different genes provide a signal and at what level they produce noise across the entire topology (Table 3). Across all the ingroup nodes, about 77% of the signal is derived from 28S and EF-1α, with 23% provided by the other genes. At the interfamilial level, COII provides no signal, whereas EF-1α and 28S provide about 85% of the signal. At the level of intrafamilial relationships, different genes provide different signal strengths in different groups. For instance, EF-1α provides a very limited signal for relationships among fleas (7.1%), although it provides more than half of the signal for relationships among the bittacids (56.5%). COII provides almost no signal for boreid relationships, but it accounts for about 20% of the signal in fleas and panorpids and 33% of the signal in bittacids. 18S provides negative support among the bittacids, but good support within the boreids. 28S appears to be the most useful individual marker as it provides roughly 40% of the signal across all ingroup nodes.
| Node partitions | Total Bremer support | Three partitioned Bremer | Percent partitioned Bremer | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 18S | 28S | EF-1α | COII | 18S | 28S | EF-1α | COII | ||
| Ingroup nodes | 1041 | 119.9 | 434.7 | 361.8 | 124.7 | 11.5 | 41.8 | 34.8 | 12.0 |
| Interfamilial nodes | 319 | 57.3 | 138.5 | 133 | −9.8 | 18.0 | 43.4 | 41.7 | −3.1 |
| Intrafamilial nodes | 722 | 62.6 | 296.2 | 228.8 | 134.5 | 8.7 | 41.0 | 31.7 | 18.6 |
| Intrafamilial (flea) | 211 | 52.5 | 101.5 | 15.0 | 42.0 | 12.1 | 48.1 | 7.1 | 19.9 |
| Intrafamilial (boreids) | 103 | 27.4 | 37.2 | 35.6 | 2.8 | 26.6 | 36.1 | 34.6 | 2.7 |
| Intrafamilial (bittacids) | 65 | −31.0 | 36.5 | 37.7 | 21.9 | −47.7 | 56.2 | 58.0 | 33.7 |
| Intrafamilial (panorpids) | 237 | 11.2 | 91.6 | 88.4 | 45.8 | 4.7 | 38.6 | 37.3 | 19.3 |
| All nodes | 1608 | 297.7 | 785.9 | 400.5 | 124 | 18.5 | 48.9 | 24.9 | 7.7 |
The combination of all these data together in a single analysis with all characters weighted equally produces a single, fully resolved topology (Fig. 5; support values in Table 4). This analysis supports a major division of Mecoptera into two clades: (Nannochoristidae (Boreidae + Siphonaptera)) and the remaining Mecoptera. The clade Siphonaptera + Boreidae is the best supported higher level relationship on the topology (Bremer support = 10; bootstrap = 60). This is congruent with earlier molecular studies which included a much smaller sample of mecopteran and flea taxa and fewer genetic markers (Whiting et al. 1997; Whiting 2001, 2002). The position of Nannochoristidae at the base of this clade is supported with less Bremer support (= 2), but a slightly higher bootstrap value (= 63). The basal placement of this family relative to other mecopteran groups accords with morphological evidence (Kristensen 1989; Willmann 1989).
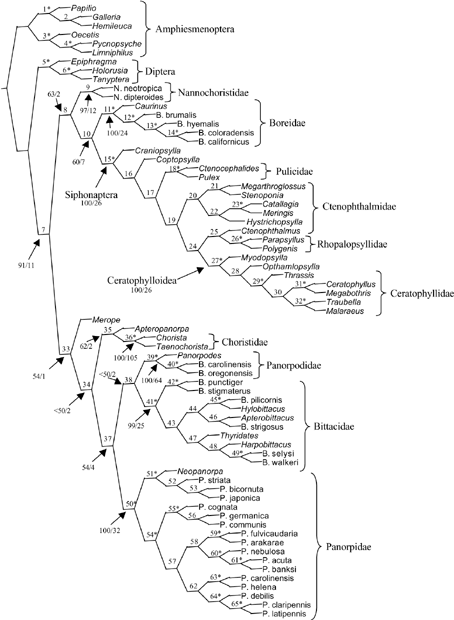
Total evidence molecular tree based on 18S + 28S + EF-1α + COII with all characters weighted equally. This analysis produces a single most parsimonious tree (L = 12 376; CI = 0.40, RI = 0.66). Nodes are numbered and Bremer and bootstrap values are given in Table 4. Nodes where bootstrap > 98 and Bremer > 10 are indicated with an asterisk. Bootstrap and Bremer values are listed for all interfamilial relationships.
| Node | Bootstrap support | Bremmer support | Partitioned Bremer | Bootstrap support | Bremer support | Partitioned Bremer | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18S | 28S | EF-1α | COII | Node | 18S | 28S | EF-1α | COII | |||||
| 1 | 100 | 83 | 33.7 | 49.6 | −2.6 | 2.3 | 34 | < 50 | 2 | 0.7 | −0.4 | −1.6 | 3.3 |
| 2 | 64 | 1 | 0.9 | −0.4 | 3.9 | −3.4 | 35 | 62 | 2 | −7.1 | −3.4 | 18.9 | −6.4 |
| 3 | 100 | 42 | 14.0 | 28.0 | 0 | 0 | 36 | 100 | 105 | 24.4 | 30.1 | 57.4 | −6.9 |
| 4 | 100 | 11 | 1.0 | 10.0 | 0 | 0 | 37 | 54 | 4 | −13.6 | 20.6 | 7.4 | −10.4 |
| 5 | 100 | 324 | 118.5 | 209.4 | 1.0 | −4.9 | 38 | < 50 | 2 | −6.6 | 7.6 | 8.4 | −7.4 |
| 6 | 100 | 106 | 9.7 | 54.6 | 36.4 | 5.3 | 39 | 100 | 64 | 16.7 | 18.6 | 16.4 | 12.3 |
| 7 | 91 | 11 | 10.2 | 5.6 | 1.9 | −6.7 | 40 | 100 | 60 | 7.4 | 19.9 | 29.4 | 3.3 |
| 8 | 63 | 2 | −1.1 | 3.6 | −1.1 | 0.6 | 41 | 99 | 25 | −7.3 | −2.4 | 19.4 | 15.3 |
| 9 | 97 | 12 | 8.2 | 8.6 | 1.9 | −6.7 | 42 | 100 | 44 | 3.4 | 10.9 | 16.6 | 13.1 |
| 10 | 60 | 7 | 0.4 | 9.6 | −6.6 | 3.6 | 43 | < 50 | 2 | −8.3 | −1.4 | 6.1 | 5.6 |
| 11 | 100 | 24 | 7.9 | 23.1 | −5.1 | −1.9 | 44 | 57 | 2 | −8.3 | −1.4 | 6.1 | 5.6 |
| 12 | 100 | 29 | 5.9 | 8.6 | 17.9 | −3.4 | 45 | 99 | 25 | −8.6 | 0.6 | 16.4 | 16.6 |
| 13 | 100 | 32 | 9.7 | 12.3 | 7.4 | 2.6 | 46 | 53 | 2 | 0.4 | −0.1 | −0.4 | 2.1 |
| 14 | 100 | 42 | 11.8 | 16.3 | 10.3 | 3.6 | 47 | 63 | 4 | −13.6 | 20.6 | 7.4 | −10.4 |
| 15 | 100 | 26 | 11.9 | 12.6 | 3.9 | −2.4 | 48 | < 50 | 2 | 0.4 | −0.1 | −0.4 | 2.1 |
| 16 | 67 | 10 | −2.1 | 7.6 | 2.9 | 1.6 | 49 | 100 | 30 | −1.3 | 16.9 | 8.6 | 5.9 |
| 17 | < 50 | 3 | −0.6 | 9.6 | −11.6 | 5.6 | 50 | 100 | 32 | 11.9 | 5.1 | 13.4 | 1.6 |
| 18 | 100 | 34 | 4.5 | 19.7 | 7.7 | 2.2 | 51 | 98 | 16 | 3.1 | 7.6 | 9.7 | −4.4 |
| 19 | < 50 | 3 | −0.6 | 8.1 | −9.1 | 4.6 | 52 | 88 | 9 | −4.0 | 5.0 | 11.0 | −3.0 |
| 20 | < 50 | 5 | 1.2 | 5.6 | 4.9 | −6.7 | 53 | 87 | 6 | 4.9 | 0.6 | 10.9 | −10.4 |
| 21 | < 50 | 3 | 5.4 | 1.6 | −1.6 | −2.4 | 54 | 100 | 26 | 2.2 | 13.6 | 15.1 | −4.8 |
| 22 | < 50 | 1 | 5.4 | 0.6 | −1.6 | −3.4 | 55 | 100 | 21 | −1.6 | 23.6 | 0.4 | −1.4 |
| 23 | 100 | 27 | 9.4 | 3.0 | 14.8 | −0.2 | 56 | < 50 | 4 | −0.3 | −2.6 | 1.1 | 5.8 |
| 24 | < 50 | 3 | 2.9 | 2.6 | −2.6 | 0.1 | 57 | 65 | 6 | 1.7 | 2.9 | −2.9 | 4.3 |
| 25 | < 50 | 2 | −1.6 | −2.4 | 0.4 | 5.6 | 58 | < 50 | 3 | −4.1 | 3.3 | 3.4 | 0.4 |
| 26 | 100 | 32 | 6.4 | 15.6 | 5.4 | 4.6 | 59 | 100 | 35 | 4.9 | 13.5 | 9.1 | 7.4 |
| 27 | 100 | 26 | 5.4 | 13.1 | 0.4 | 7.1 | 60 | 100 | 50 | 3.7 | 8.2 | 12.5 | 25.6 |
| 28 | < 50 | 3 | 2.9 | 2.6 | −2.6 | 0.1 | 61 | 100 | 11 | 1.0 | 6.0 | 1.0 | 3.0 |
| 29 | 99 | 17 | 4.4 | 2.1 | 3.9 | 6.6 | 62 | 84 | 5 | 0.0 | 4.0 | 0.0 | 1.0 |
| 30 | 56 | 3 | 2.9 | 2.6 | −2.6 | 0.1 | 63 | 100 | 18 | −0.3 | 2.9 | 6.1 | 9.3 |
| 31 | 100 | 17 | 4.9 | 5.6 | −1.1 | 7.6 | 64 | 99 | 9 | 0.0 | 0.0 | 5.0 | 4.0 |
| 32 | 100 | 22 | 1.7 | 3.9 | 7.4 | 8.9 | 65 | 100 | 18 | 0.0 | 3.0 | 6.0 | 9.0 |
| 33 | 54 | 1 | 0.7 | −0.4 | −1.6 | 2.3 | |||||||
A sister group relationship between Boreidae and Siphonaptera is also supported by morphological evidence. The process of resilin secretion in the flea (pleural arch) and Boreus (wing base) is similar, and different from that of the locust and dragonfly (Rothschild 1975; Schlein 1980). The unusual proventricular spines in fleas and boreids are morphologically similar (Richards & Richards 1969). Both groups have multiple sex chromosomes (Bayreuther & Brauning 1971) and also have eyes in a ‘skeletal socket’ (Schlein 1980). Boudreaux (1979) considered the above characters as probable convergences, and favoured a placement of Siphonaptera as sister group to Diptera, andByers (1996) presented arguments for a close association of fleas with flies. Nonetheless, the most convincing morphological evidence comes from recent research on ovarioles, which demonstrates that boreid ovarioles are fundamentally different from those in other Mecoptera, but similar to those found in fleas. Mecoptera possess polytrophic–meroistic ovarioles, whereas the ovarioles in Boreus are devoid of nurse cells and therefore panoistic (Bilinski et al. 1998). Fleas and boreids share the following ovariole characteristics: (i) secondary loss of nurse cells; (ii) completion of initial stages of oogenesis during postembryonic development; (iii) occurrence of rDNA amplification and resulting appearance of multiple nucleoli; (iv) differentiation of the late previtellogenic ooplasm into two clearly recognizable regions; and (v) presence of accumulations of membrane-free, clathrin-like cages (Bilinski et al. 1998). The combination of morphological with molecular data provides a compelling argument for a sister group relationship between Boreidae and Siphonaptera.
The second major clade supported by the combined data includes the remainder of Mecoptera, with Meropeidae as the basal-most member of this clade. There were no sequences included from Eomeropidae, and so it is not clear whether ‘Protomecoptera’sensuKaltenbach (1978) is supported. These data support a sister group relationship between Apteropanorpidae and Choristidae. The combined analysis favours a sister group relationship between Panorpidae and Bittacidae, and this finding contradicts results from previous morphological analyses which favour Panorpidae + Panorpodidae, although the position of Bittacidae has always been open to question. It is interesting that the Panorpidae + Panorpodidae clade, which is thought to be well supported via morphological data (Willman 1987), was never well supported in any of the gene partitions. Three gene partitions directly contradict Panorpidae + Panorpodidae, and, in the fourth (18S rDNA), the relationship is poorly supported. Likewise, the Bittacidae + Panorpidae relationship in the combined analysis is poorly supported, and Bittacidae are placed with different clades for every gene partition in this analysis. These observations suggest that further data are needed to establish a robust placement for Bittacidae.
In contrast to the marginally supported interfamilial relationships, the monophyly of every mecopteran family is very well supported (minimum bootstrap = 97; minimum Bremer = 12), as are many of the generic and species group relationships within Mecoptera and Siphonaptera. Within Siphonaptera, the families Ceratophyllidae, Rhopalopsyllidae and Pulicidae, and the superfamilial group Ceratophylloidea, are well supported, but the data suggest that Ctenophthalmidae is paraphyletic. This analysis supports Craniopsylla as the most basal flea taxon and Caurinus as the most basal boreid. Although there has been no previous formal analysis of phylogenetic relationships within Panorpidae, the species group designations suggested by Carpenter (1931) and Issiki (1935) are supported in this analysis, including the Japonica group (P. striata, bicornuta and japonica), the Communis group (P. cognata, germanica and communis), the Fulvicaudaria group (P. fulvicaudaria and arakarae), the Nebulosa group (P. nebulosa, acuta and banksi), the Helena group (P. carolinensis and helena) and the Claripennis group (P. claripennis and latipennis). The genus Panorpa is paraphyletic, as Neopanorpa is placed as sister taxon to the Japonica species group. Likewise, within Bittacidae, the genus Bittacus is grossly paraphyletic with regard to the other bittacid genera. The fact that these two genera are paraphyletic is not particularly surprising as both are catch-all genera that include a wide range of species from throughout the world. Within Panorpodidae, the two Brachypanorpa species are sister taxa as expected from morphology.
These data suggest that Mecoptera, as currently constituted, is a paraphyletic assemblage. While it seems certain that Boreidae and Siphonaptera are sister groups, their placement relative to the other Mecoptera is not as well supported by the data. Likewise, while it seems clear that Nannochoristidae should occupy a basal position, it is not clear whether it is sister group to the flea + boreid clade or sister to the remainder of Mecoptera. Additional data in the form of increased taxon sampling for the molecular data and a coded morphological matrix are needed to provide a more robust estimate of mecopteran and flea relationships.
Acknowledgements
I thank Alison Whiting and Paige Humphreys for assistance in generating the sequence data. Thanks go to many researchers who provided taxa used in this analysis, including H. Aberlene, T. Hoernschemeyer, J. McHugh, R. Meier, B. Misof, H. Ogai, N. Penny, L. Russell, B. Wiegmann, A. Whittington and D. Yeates. Special thanks are extended to G. W. Byers for his extensive knowledge of Mecoptera, for critical assistance and for encouragement in all stages of this project. M. W. Hastriter provided the flea specimens and identifications for this study. This work was supported by NSF grants DEB-9615269 and DEB-9806349, and NSF CAREER award DEB-9983195.
Appendix
| Family | Name | 18S | 28S | EF-1 | COII |
|---|---|---|---|---|---|
| Papilionidae | Papilio troilus L. 1758 | AF286299 | AF423920 | AF423810 | AF423981 |
| Pyralidae | Galleria melonella (L. 1758) | AF286298 | AF423921 | AF423811 | AF423982 |
| Saturniidae | Hemileuca sp. Walker 1855 | AF286273 | AF423922 | AF423812 | AF423983 |
| Leptoceridae | Oecetis avara Banks 1895 | AF286300 | AF423917 | AF423815 | AF423986 |
| Limniphilidae | Pycnopsyche lepida (Hagen 1861) | AF286292 | AF423923 | AF423813 | AF423984 |
| Limniphilidae | Limnephilus sp. Leach 1815 | AF286291 | AF338267 | AF423814 | AF423985 |
| Tipulidae | Epiphragma fasciapenne (Say 1823) | AF286294 | AF423919 | AF423808 | AF423979 |
| Tipulidae | Holorusia rubiginosa Loew 1863 | AF423778 | AF423924 | AF423809 | AF423980 |
| Tipulidae | Tanyptera dorsalis (Walker 1848) | AF286295 | AF423918 | AF423807 | AF423978 |
| Nannochoristidae | Nannochorista neotropica Navas 1928 | AF334799 | AF338261 | AF423848 | AF424018 |
| Nannochoristidae | Nannochorista dipteroides Tillyard 1917 | AF334796 | AF338262 | AF423849 | AF424019 |
| Boreidae | Caurinus dectes Russell 1979 | AF286288 | AF423937 | AF423830 | AF424001 |
| Boreidae | Boreus brumalis Fitch 1847 | AF423883 | AF423936 | AF423828 | AF423999 |
| Boreidae | Boreus hyemalis (L. 1767) | AF423882 | AF423935 | AF423827 | AF423998 |
| Boreidae | Boreus colouradensis Byers 1955 | AF286285 | AF423934 | AF423826 | AF423997 |
| Boreidae | Boreus californicus Packard 1870 | AF334795 | AF338257 | AF423829 | AF424000 |
| Meropeidae | Merope tuber Newman 1838 | AF286287 | AF338260 | AF423847 | AF424017 |
| Apteropanorpidae | Apteropanorpa evansi Byers and Yeates 1999 | AF286284 | AF423925 | AF423816 | AF423987 |
| Choristidae | Chorista australis Klug 1838 | AF286289 | AF423943 | AF423836 | AF424007 |
| Choristidae | Taeniochorista pallida Esben-Petersen 1914 | AF423889 | AF423944 | AF423837 | AF424008 |
| Panorpodidae | Brachypanorpa carolinensis Banks 1905 | AF286296 | AF423971 | AF423867 | AF424037 |
| Panorpodidae | Brachypanorpa oregonensis (McLachlan 1881) | AF423912 | AF423972 | AF423868 | AF424038 |
| Panorpodidae | Panorpodes pulcher Issiki 1927 | AF423913 | AF423973 | AF423869 | AF424039 |
| Bittacidae | Apterobittacus apterus (McLachlan 1871) | AF423875 | AF423926 | AF423817 | AF423988 |
| Bittacidae | Bittacus pillicornis Westwood 1846 | AF334800 | AF338256 | AF423822 | AF423993 |
| Bittacidae | Bittacus punctiger Westwood 1846 | AF423876 | AF423927 | AF423818 | AF423989 |
| Bittacidae | Bittacus selysi Esben-Petersen 1917 | AF423878 | AF423929 | AF423820 | AF423991 |
| Bittacidae | Bittacus stigmaterus Say 1823 | AF423881 | AF423932 | AF423824 | AF423995 |
| Bittacidae | Bittacus strigosus Hagen 1861 | AF286290 | AF423933 | AF423825 | AF423996 |
| Bittacidae | Bittacus walkeri Esben-Petersen 1915 | AF423879 | AF423930 | AF423821 | AF423992 |
| Bittacidae | Harpobittacus australis rubipes Riek 1954 | AF423877 | AF423928 | AF423819 | AF423990 |
| Bittacidae | Hylobittacus apicalis (Hagen 1861) | AF423880 | AF423931 | AF423823 | AF423994 |
| Panorpidae | Neopanorpa harmandi (Navas 1908) | AF423903 | AF423961 | AF423856 | AF424027 |
| Panorpidae | Panorpa acuta Carpenter 1931 | AF423908 | AF423967 | AF423863 | AF424033 |
| Panorpidae | Panorpa arakavae Miyake 1913 | AF423901 | AF423959 | AF423854 | AF424025 |
| Panorpidae | Panorpa banksi Hine 1901 | AF423909 | AF423968 | AF423864 | AF424034 |
| Panorpidae | Panorpa bicornuta McLachlan 1887 | AF423902 | AF423960 | AF423855 | AF424026 |
| Panorpidae | Panorpa carolinensis Banks 1905 | AF423898 | AF423955 | AF423852 | AF424022 |
| Panorpidae | Panorpa claripennis Hine 1901 | AF423904 | AF423962 | AF423858 | AF424028 |
| Panorpidae | Panorpa cognata Rambur 1842 | AF423897 | AF423954 | AF423851 | AF424021 |
| Panorpidae | Panorpa communis L. 1758 | AF423900 | AF423957 | AF423857 | AF424024 |
| Panorpidae | Panorpa debilis Westwood 1846 | AF423899 | AF423956 | AF423853 | AF424023 |
| Panorpidae | Panorpa fluvicaudaria Miyake 1910 | AF423896 | AF423953 | AF423850 | AF424020 |
| Panorpidae | Panorpa germanica L. 1758 | AF423907 | AF423965 | AF423862 | AF424032 |
| Panorpidae | Panorpa helena Byers 1962 | AF334798 | AF338264 | AF423859 | AF424029 |
| Panorpidae | Panorpa japonica Thunberg 1784 | AF423910 | AF423969 | AF423865 | AF424035 |
| Panorpidae | Panorpa latipennis Hine 1901 | AF423906 | AF423964 | AF423861 | AF424031 |
| Panorpidae | Panorpa nebulosa Westwood 1846 | AF423905 | AF423963 | AF423860 | AF424030 |
| Panorpidae | Panorpa striata Miyake 1908 | AF423911 | AF423970 | AF423866 | AF424036 |
| Stephanocircidae | Craneopsylla minerva wolffheuglia (Rothschild 1909) | AF286286 | AF338266 | AF423874 | AF424044 |
| Coptopsyllidae | Coptopsylla africana Wagner 1932 | AF286275 | AF423945 | AF423838 | AF424009 |
| Pulicidae | Ctenocephalides canis (Curtis 1826) | AF423914 | AF423974 | AF423870 | AF424040 |
| Pulicidae | Pulex irritans L. 1758 | AF423915 | AF423975 | AF423871 | AF424041 |
| Ctenophthalmidae | Megarthroglossus divisus (Baker 1898) | AF286276 | AF338258 | AF423839 | AF424010 |
| Ctenophthalmidae | Stenoponia americana (Baker 1899) | AF423893 | AF423949 | AF423843 | AF424014 |
| Ctenophthalmidae | Catallagia sp. | AF423890 | AF423946 | AF423840 | AF424011 |
| Ctenophthalmidae | Meringis hubbardi Kohls 1938 | AF423891 | AF423947 | AF423841 | AF424012 |
| Hystrichopsyllidae | Hystrichopsylla talpae talpae (Curtis 1826) | AF286281 | AF423950 | AF423844 | AF424015 |
| Ctenophthalmidae | Ctenopthalmus p. pseudagyrtes Baker 1904 | AF423892 | AF423948 | AF423842 | AF424013 |
| Rhopalopsyllidae | Parapsyllus magellanicus largificus Smit 1984 | AF423916 | AF423976 | AF423872 | AF424042 |
| Rhopalopsyllidae | Polygenis pradoi (Wagner 1937) | AF286277 | AF423977 | AF423873 | AF424043 |
| Ischnopsyllidae | Myodopsylla gentilis Jordan & Rothschild 1921 | AF423894 | AF423951 | AF423845 | |
| Leptopsyllidae | Opthalmopsylla volgensis palestinica Smit 1960 | AF423895 | AF423952 | AF423846 | AF424016 |
| Ceratophyllidae | Thrassis bacchi gladiolus (Jordan 1925) | AF423886 | AF423940 | AF423833 | AF424004 |
| Ceratophyllidae | Ceratophyllus petrochelidoni Wagner 1936 | AF423888 | AF423942 | AF423835 | AF424006 |
| Ceratophyllidae | Megabothris calcarifer (Wagner 1913) | AF423887 | AF423941 | AF423834 | AF424005 |
| Ceratophyllidae | Traubella grundmanni Egoscue 1989 | AF423884 | AF423938 | AF423831 | AF424002 |
| Ceratophyllidae | Malaraeus sinomus (Jordan 1925) | AF423885 | AF423939 | AF423832 | AF424003 |




