Effect of Genetic Variation in the Human Fatty Acid Synthase Gene (FASN) on Obesity and Fat Depot-Specific mRNA Expression
Abstract
Inhibition of fatty acid synthase (FASN) induces a rapid decline in fat stores in mice, suggesting a role for this enzyme in energy homeostasis. To investigate the potential role of FASN in the pathophysiology of human obesity, the FASN gene was sequenced in 48 German whites. Thirty-five single-nucleotide polymorphisms (SNPs) were identified. Eight SNPs representative for their linkage disequilibrium groups and the Val1483Ile (rs2228305) substitution were genotyped for subsequent association analyses in 1,311 adults from Germany. Further, the tagging SNPs were genotyped also in German childhood cohorts (738 schoolchildren, 205 obese children). Effects of genetic variation on FASN mRNA expression in visceral and subcutaneous adipose tissue from a subgroup of 172 subjects were analyzed. Several polymorphisms in the FASN (rs62078748, rs2229422, rs2229425, and rs17848939) were nominally associated with obesity in case–control studies including 446 obese subjects (BMI ≥30 kg/m2) and 389 lean controls (BMI ≤25 kg/m2) (adjusted P < 0.05). The strongest significant effect was found for rs2229422 (P = 1.3 × 10−5 adjusted for age, sex, type 2 diabetes status), which was supported by associations with BMI, waist-to-hip ratio (WHR), fasting plasma insulin and glucose infusion rate (adjusted P < 0.05). Subjects with the Val1483Ile substitution appeared to be protected against obesity. In addition, rs17848939 was nominally significantly associated with the ratio of visceral/subcutaneous FASN mRNA expression (adjusted P = 0.04). No effect of genetic variation in FASN on obesity was found in children. In conclusion, our data indicate a role of FASN genetic variation in susceptibility to obesity in adults.
Introduction
Obesity is a significant risk factor for many life-threatening diseases, and its global impact on health is enormous (1). Although the etiology of obesity is poorly understood, this disease has both environmental and genetic components.
It has been postulated that increased expression and activity of the lipogenic pathways in adipose tissue could play a role in obesity development (2). The central enzyme in the pathway of de novo lipogenesis, fatty acid synthase (FASN), catalyses all steps in the conversion of malonyl CoA to palmitate (3). Although reports on genetic variation in human FASN are rare, there are a number of recent studies from rodents indicating that FASN may be involved in obesity through regulation of feeding behavior (4,5,6). Treatment of mice with FASN inhibitors (cerulenin, C75) results in reduced food intake and substantial body weight loss (4). The FASN inhibitor, C75, acts both centrally to reduce food intake and peripherally to increase fatty acid oxidation, leading to rapid and profound weight loss, loss of adipose mass, and resolution of fatty liver in diet-induced obese mice (7).
The human FASN maps on chromosome 17q25 (complement strand) within a region of linkage for body fat content in Pima Indians (8). Sequence analysis of FASN in this genetically and environmentally homogeneous population of Southern Arizona characterized by obesity and high prevalence of type 2 diabetes mellitus (T2DM) (9,10), revealed a novel Val1483Ile polymorphism that was shown to be associated with percentage of body fat and 24-h substrate oxidation rates measured in a respiratory chamber (11). It has been suggested that the Val1483Ile substitution in FASN is protective against obesity, an effect possibly explained by the role of this gene in the regulation of substrate oxidation (11). The Pima data have recently been supported by studies in white children, reporting a sex-specific protective effect of the Val1483Ile variant for obesity as well as its beneficial effect on lipid profile in obese boys (12). However, comprehensive genetic analyses of FASN such as the one reported in Pima Indians are still lacking in white populations.
Recently, we examined whether FASN mRNA expression in visceral and subcutaneous fat correlates with anthropometric and metabolic parameters in subjects with a wide range of obesity, body fat distribution, insulin sensitivity, and glucose tolerance (13). Our data clearly demonstrated significant association of increased FASN expression in adipose tissue not only with obesity, but also with T2DM and insulin resistance, thus indicating a possible role of lipogenic pathways in the development of human obesity and/or T2DM (13).
Therefore, in the present study we sequenced the gene in subjects from Eastern Germany and assessed the effects of identified polymorphisms on human obesity/T2DM and pathophysiologically relevant traits. We further investigated whether genetic variation within the FASN might be responsible for changes in FASN expression in adipose tissue.
Experimental subjects
Leipzig adult cohort. In total, 640 patients with T2DM and 671 healthy subjects were recruited at the University Hospital in Leipzig, Germany. BMI was calculated as weight divided by squared height. Waist and hip circumferences were measured and waist-to-hip ratio (WHR) was calculated. The healthy subjects included 246 males and 425 females (mean age 46.5 ± 14.4 years, mean BMI 27.5 ± 5.4 kg/m2) and patients with T2DM included 301 males and 339 females (mean age 63.3 ± 11.3 years, mean BMI 29.8 ± 5.2 kg/m2; the data represent mean ± s.d.). In addition oral glucose tolerance test and fasting plasma insulin measurements were performed in all nondiabetic subjects. The oral glucose tolerance test was performed according to the criteria of the American Diabetes Association (14). The test was carried out after an overnight fast with 75 g standardized glucose solution (Glucodex Solution 75 g; Merieux, Montreal, Canada). Venous blood samples were taken at 0, 60, and 120 min for measurements of plasma glucose concentrations. 196 out of 671 subjects had impaired glucose tolerance. As impaired glucose tolerance is a T2DM predicting factor, only remaining 502 subjects with normal glucose tolerance were included as healthy controls in T2DM association study.
In a subgroup of 380 nondiabetic subjects, body fat content and insulin sensitivity were measured. Percentage body fat was measured by dual-energy X-ray absorptiometry. Insulin sensitivity was assessed with the euglycemic–hyperinsulinemic clamp method as previously described (15,16).
In addition, paired samples of visceral and subcutaneous adipose tissue and DNA were obtained from a subgroup of 172 white men (N = 84) and women (N = 88), who underwent open abdominal surgery for gastric banding, cholecystectomy, weight reduction surgery, abdominal injuries, or explorative laparotomy. The age ranged from 16 to 99 years and BMI from 20.8 to 54.1 kg/m2. Thirty-eight subjects had T2DM and 15 subjects had impaired glucose tolerance. All subjects had a stable weight with no fluctuations of >2 percent of the body weight for at least 3 months before surgery. Patients with severe conditions including generalized inflammation or end stage malignant diseases were excluded from the study. Samples of visceral and subcutaneous adipose tissue were immediately frozen in liquid nitrogen after explantation.
The white status of study subjects was self-reported. All studies were approved by the ethics committee of the University of Leipzig and all subjects gave written informed consent before taking part in the study.
Childhood cohorts
Leipzig Schoolchildren cohort: This cohort is part of the Leipzig Schoolchildren Project that investigated anthropometric and clinical parameters in 2,675 children aged 6 to 17 years from 1999–2000 (17). DNA was available from 738 children (358 boys and 380 girls; mean age 12 ± 3 years; mean BMI-SDS 0.09 ± 0.98). The BMI was standardized referring to national reference data (18) and children with a BMI ≥1.88 SDS (= 97th percentile) were classified obese. A total of 508 children and adolescents with BMI between −1.0 SDS and 1.0 SDS (243 boys and 265 girls; mean age 12 ± 3 years; mean BMI-SDS −0.05 ± 0.54) were selected from the 738 schoolchildren to serve as healthy normal weight control group to Leipzig obese children cohort.
Leipzig obese children cohort: A total of 205 white children and adolescents (97 boys and 108 girls; 12 ± 4 years; BMI-SDS 2.71 ± 0.58) were recruited from the obesity clinic at the University Hospital for Children & Adolescents, Leipzig, Germany. All obese children had a detailed metabolic work up including an oral glucose tolerance test as described in detail elsewhere (19).
The studies were approved by the ethics committee of the University of Leipzig.
Methods and Procedures
Analysis of human FASN gene expression
Total RNA was isolated from paired subcutaneous and visceral adipose tissue samples using TRIzol (Life Technologies, Grand Island, NY), 1 µg RNA was reverse transcribed with standard reagents (Life Technologies). From each reverse transcription–PCR, 2 µl was amplified in a 26-µl PCR using the Brilliant SYBR Green QPCR Core Reagent Kit from Stratagene (La Jolla, CA) according to the manufacturer's instructions. Samples were incubated in the ABI PRISM 7000 sequence detector for an initial denaturation at 95 °C for 10 min, followed by 40 PCR cycles, each cycle consisting of 95 °C for 15 s, 60 °C for 1 min, and 72 °C for 1 min. The following primers were used: human FASN (accession no. NM_004104) 5′-CGCGTGGCCGGCTACTCCTAC-3′ (sense) and 5′-CGGCTGCCACACGCTCCTCT-3′ (antisense). Human FASN mRNA expression was calculated relative to the mRNA expression of 18S rRNA, determined by a premixed assay on demand for human 18S rRNA (PE Biosystems, Foster City, CA). Amplification of specific transcripts was confirmed by melting curve profiles (cooling the sample to 68 °C and heating slowly to 95 °C with measurement of fluorescence) at the end of each PCR. The specificity of the PCR was further verified by subjecting the amplification products to agarose gel electrophoresis.
Sequencing of the FASN
To identify genetic variants, all 43 exons, including intron/exon splicing sites (the total length of FASN mRNA is 8,481 bp, NM_004104), 2,280 bp upstream of the first translation initiation site and 840 bp of the 3′ untranslated region were sequenced in DNA samples from 48 nonrelated German subjects (24 obese and 24 with normal weight). Sequencing was performed using the Big Dye Terminator (Applied Biosystems, Foster City, CA) on an automated DNA capillary sequencer (ABI PRISM 3100 Avant; Applied Biosystems). Sequence information for all oligonucleotide primers used for variant screening is available upon request.
Genotyping of FASN SNPs
Genotyping of selected single-nucleotide polymorphisms (SNPs) in all study subjects was done using the TaqMan assay (Applied Biosystems) for the rs17848935, rs4485435, rs2229422, rs4246445, rs2229425, rs2228309, rs62078748, rs2228305 variants and by restriction fragment length polymorphism technique for the rs17848939 variant. The TaqMan genotyping reaction was amplified on a GeneAmp PCR system 9700 (95 °C for 15 min, and 95 °C for 15 s, and 60 °C for 1 min, for 40 cycles) and fluorescence was detected on an ABI PRISM 7700 sequence detector (Applied Biosystems). The restriction fragment length polymorphism genotypes were determined by PCR amplification of the respective fragments of the FASN on a GeneAmp PCR system 9700 (94 °C for 3 min, and 94 °C for 30 s, and 66 °C for 45 s, 72 °C for 45 s, for 35 cycles, and 72 °C for 10 min) and subsequent digestion with the NotI restriction enzyme and size fractionation and visualization by electrophoresis. To assess genotyping reproducibility, a random ∼10% selection of the sample was regenotyped for each SNP; all genotypes matched initial designated genotypes. Moreover, the DNA samples selected for sequencing were regenotyped by TaqMan technique, as they were also included in the association analyses. The genotypes matched completely between the two techniques.
Statistical analyses
Prior to statistical analysis, non-normally distributed parameters (glucose infusion rate, 2-h plasma glucose, fasting plasma insulin, visceral and subcutaneous FASN mRNA expression) were logarithmically transformed to approximate a Gaussian distribution. Differences in genotype frequencies were compared using logistic regression models between the obese cases and lean controls as well as between diabetic cases and healthy controls with normal glucose tolerance. Multivariate linear relationships were assessed by generalized linear model analyses. In the additive model, homozygotes for the major allele (MM), heterozygotes (Mm) and homozygotes for the minor allele (mm) were coded to a continuous numeric variable for genotype (as 0, 1, and 2, respectively). A dominant model was defined as contrasting genotypic groups MM+Mm vs. mm, and the recessive model was defined as contrasting genotypic groups MM vs. Mm+mm. In haplotype analyses, groups of subjects carrying 2, 1, or 0 copies of the haplotype were compared. To estimate haplotypes we used the PHASE version 2.1 software (20,21). The phase reconstruction method considers the unknown haplotypes as unobserved random quantities and aims to evaluate their conditional distribution in light of the genotype data. Statistical analyses were performed using the SPSS software package (SPSS, Chicago, IL) and the statistical analysis system of the SAS Institute (Cary, NC). All data are presented without correcting for multiple testing. Bonferroni corrections for multiple testing would require a P value of 0.006 for analyses of associations in the present case–control studies and P value of 0.007 for the association analyses with quantitative traits to consider the findings significant. P values < 0.05 but >0.006 were therefore considered to be of nominal statistical significance.
Results
Genetic variation in the FASN
The FASN gene (all 43 Exons with Intron/Exon splicing sites, 2 kb in the 5′ region and 840 bp of the 3′ untranslated region) was sequenced in 48 white DNA samples (24 obese and 24 lean subjects) to identify variation. Thirty-five SNPs including 14 novel variants were identified: 12 SNPs were in the 5′ region, 14 SNPs in exons, 7 SNPs in introns and 2 SNPs in the 3′ untranslated region (Table 1). The genotype distributions for all SNPs were consistent with Hardy–Weinberg equilibrium (P > 0.05), except for rs1140616 which was therefore excluded from further analyses. Linkage disequilibrium (r2 and D-prime (D′)) was calculated among the 16 common variants (minor allele frequency (MAF) >0.05) using the EMLD statistical program (https:epi.mdanderson.orgqhuangSoftwarepub.htm) (Supplementary Table S1 online). Based on LD between these common variants, eight groups were defined by the Tagger software (22) (Supplementary Table S2 online). Because SNPs in high genotypic concordance would provide the same information for association studies, one representative variant for each LD-group was selected and genotyped in all subjects for association analyses (Supplementary Table S2 online). In addition we genotyped the Val1483Ile (rs2228305) variant (MAF = 4%) in the adult cohort, which has previously been reported to be associated with obesity in different ethnicities (11,12).
 |
Moreover, using the PHASE version 2.1 software (20,21), we identified five common haplotypes (frequency of each haplotype ≥0.05) among the eight different SNPs genotyped in all subjects. These five haplotypes, [GCTCTGCC], [GTCCTACT], [GTCGTACC], [ACCCCGCC], and [ACCCCGTC] accounted for >80% of the all observed haplotypes, where haplotypes are defined by the composition of alleles at each SNP in following order: [rs62078748]-[rs2228309]-[rs17848935]-[rs4485435]-[rs2229422]-[rs17848939]-[rs2229425]-[rs4246445].
Association studies in adults
Genetic variants in the FASN and obesity. Several representative SNPs were associated with obesity under logistic regression analysis in a case–control designed study including 446 obese subjects (BMI ≥30 kg/m2, mean age: 59 ± 12 years, mean BMI: 34.6 ± 4.5 kg/m2) and 389 lean controls (BMI ≤25 kg/m2, 48 ± 18 years, 23.4 ± 1.4 kg/m2) (Table 2). Including age, sex, and T2DM status in the analyses, frequency of minor alleles for the SNPs rs62078748, rs2229422, and rs2229425 was significantly higher in the obese group compared with lean controls (P values ranging from 0.001 to 1.3 × 10−5). This conferred a significantly higher risk of obesity in additive mode of inheritance ranging from odds ratio = 1.51 (95% confidence interval (1.19; 1.92)) for the rs62078748 to 1.85 (1.35; 2.56) for the rs2229425. In contrast, the minor allele for rs17848939 was nominally significantly more frequent in the lean group (P = 0.01; in additive mode of inheritance after adjusting for age, sex, and T2DM). The analyses were performed also without subjects with T2DM; the SNP allele frequencies in the obese group remained unchanged. However, even though we attempted to eliminate some redundancy in the association testing by selecting only one SNP per each defined LD-group, the tagging SNPs seem to be still correlated (e.g., between rs62078748 and rs2229422 D′ = 0.86 and r2 = 0.63; Supplementary Table S1 online). As it is possible that a single variant could explain all the observed association, all tests were re-analyzed conditional on the strongest association found for rs2229422. After including rs2229422 in the analyses, none of the remaining SNPs showed significant association with obesity.
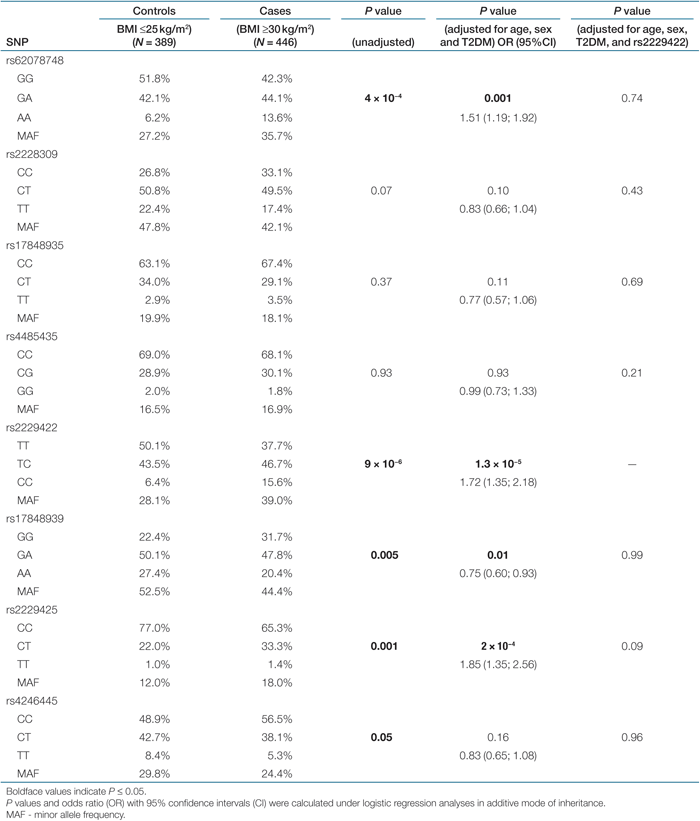 |
Consistent with the SNP data, two haplotypes were significantly associated with obesity. The frequency of the haplotype [ACCCCGCC] was significantly increased in the obese group (13.9% vs. 10.0% in the lean subjects) whereas the haplotype [GTCCTACT] showed increased frequency in lean controls (25.3% vs. 19.4% in the obese group (P = 0.003 and 0.002, respectively; in additive mode of inheritance after adjusting for age and sex). PHASE software provides a list of the best haplotype guess for each individual. To avoid misinterpreting the association results between haplotypes and phenotypic traits, we performed the analyses also in a dataset excluding all individuals where the phase probability at least at one site was <0.8. This eliminated all subjects from the analyses who may have influenced the analyses by including their uncertain haplotypes. The results of analyses with the new dataset remained materially unchanged. The P value for association of the [ACCCCGCC] haplotype with obesity changed from 0.003 to 0.009 and remained the same for the [GTCCTACT] haplotype (P = 0.002).
Genetic variants in the FASN and type 2 diabetes. In the present study, five out of eight representative SNPs were nominally significantly associated with T2DM in 640 cases and 502 healthy controls (with normal glucose tolerance) under logistic regression analysis (all P < 0.05 in additive mode of inheritance; Supplementary Table S3 online). However, after adjusting the analyses for age, sex, and BMI, only rs2228309 remained moderately associated with T2DM (P < 0.05; Supplementary Table S3 online). Consistent with these data, none of the five common haplotypes showed significant differences in frequencies between diabetic and nondiabetic subjects (all P > 0.05; data not shown).
Genetic variants in the FASN and parameters of obesity and glucose metabolism. In the cohort of 671 white subjects without T2DM, several polymorphisms showed relationship to quantitative parameters of obesity and glucose metabolism. As the strongest effect in the obesity case–control study was observed for rs2229422, we focused our analyses on this SNP. The rs2229422 C allele was significantly associated with higher BMI and higher WHR (after adjusting for age and sex; Supplementary Table S4 online). Moreover rs2229422 was nominally associated with fasting plasma insulin (adjusted for age, sex, and BMI) (Supplementary Table S4 online). Finally, the C allele was associated with lower glucose infusion rate assessed by euglycemic–hyperinsulinemic clamp (adjusted for age, sex, and BMI) (Supplementary Table S4 online). The effect of the SNP on obesity-related traits was reflected in the haplotype analysis as well, which however did not show any associations beyond those observed for the SNP (data not shown).
Val1483Ile genotype protects against obesity. In the present study, we also attempted to replicate the previously described relationship of the Val1483Ile substitution on obesity. In the entire cohort, Ile-allele carriers had nominally significantly lower BMI than noncarriers (1; P = 0.04 after adjusting for age and sex in additive mode of inheritance). After gender stratification, the association with BMI was only seen in male subjects (1). Also in the case–control study, an association with obesity was seen in males but not in females (adjusted P = 0.04 vs. P = 0.86 in additive mode of inheritance) (Table 3). Based on this, we tested a possible gender specific effect for the other investigated SNPs. Unlike for Val1483Ile, we did not observe gender differences in effects of the other SNPs on obesity and related traits. All P values in gender strata were consistent with those observed in the total cohort (data not shown).
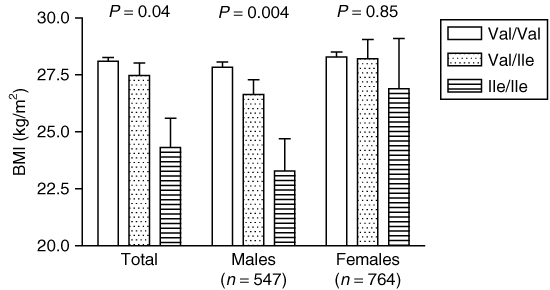
Association of the Val1483Ile (rs2228305) variant with BMI. The data are presented as mean ± s.e.m. P values were calculated after adjusting for age (and for sex in the entire cohort) in an additive mode of inheritance.
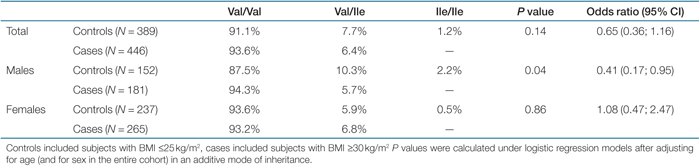 |
Although both the Val1483Ile and the rs2229422 are associated with obesity, they are not in strong linkage disequilibrium (D′ = 0.76 and r2 = 0.01) and thus, their effects seem to be independent of each other.
Association studies in children. In a case–control study including 508 lean and 205 obese children, no significant association of the FASN SNPs was found (P > 0.05 after adjusting for age, sex, pubertal stage, and height). In line with this, the FASN variants did not associate with measures of obesity (BMI, WHR) in either schoolchildren (N = 738) or obesity cohort (N = 205) (adjusted P > 0.05; data not shown).
Association of genetic variants in the FASN with fat depot-specific FASN mRNA expression. No association of the eight representative SNPs or derived haplotypes with FASN mRNA levels in either visceral or subcutaneous adipose tissue was found in 172 individuals with visceral and subcutaneous adipose tissue biopsies (all P > 0.05 after adjusting for age, sex, and BMI). To express FASN mRNA expression in visceral relative to that in subcutaneous fat, we calculated the visceral/subcutaneous FASN mRNA ratio. The rs17848939 was nominally significantly associated with the visceral to subcutaneous (visc/sc) FASN mRNA expression ratio (P = 0.04 and 0.02 in additive and recessive mode respectively, after adjusting for age, sex, and BMI) (Table 4). We also performed statistical analyses excluding subjects with impaired glucose tolerance and T2DM, but the results remained unchanged (data not shown).
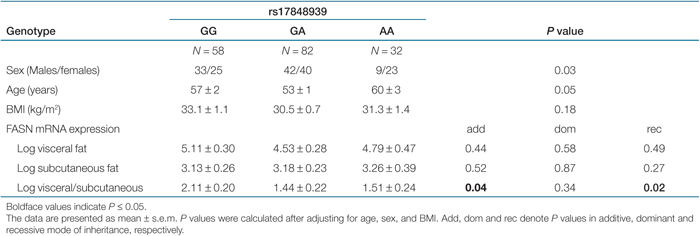 |
Discussion
The FASN has been comprehensively investigated in Pima Indians (11), but such studies are still missing in European populations. In the present study, we therefore re-sequenced FASN in subjects from Eastern Germany and investigated identified polymorphisms in subsequent association analyses. In addition, we also evaluated the role of genetic variation in the FASN on its mRNA expression in visceral and subcutaneous adipose tissue. The key findings of the present study are: (i) Genetic variants in the FASN are significantly associated with human obesity and its related traits in adult Germans and (ii) Support of previously reported sex-specific effects of the Val1483Ile variant on obesity.
In the present study, 35 genetic variants were identified by sequencing the FASN. Sixteen variants had MAF >5% and fell into eight LD groups. Association analyses of representative variants for each of the LD groups showed several polymorphisms (rs62078748, rs2229422, rs17848939, rs2229425) associated with obesity in case–control studies. Conditional analysis revealed rs2229422 to be the strongest obesity associated FASN genetic variant which was further supported through analyses in the cohort of white subjects without T2DM. The polymorphism showed relationships to quantitative measures of obesity such as BMI and WHR. Although the rs2229422 showed nominally significant associations with T2DM in the case–control study, the effect did not withstand corrections for BMI suggesting that the effect of the SNP on T2DM was mediated through obesity. None of the investigated polymorphisms with MAF >5% has an evident functional relevance for the FASN protein i.e., no SNP causes amino acid substitution in the protein or results in a new frameshift. We therefore further evaluated whether FASN genetic variants might affect the mRNA expression in visceral and subcutaneous fat and thus explain the relationship with metabolic traits related to obesity and T2DM. Recent studies carried out by our group showed that FASN mRNA expression in adipose tissue was related to measures of obesity, insulin sensitivity, and glucose tolerance (13). Furthermore, there was increased FASN mRNA expression in visceral as compared to subcutaneous adipose tissue in lean healthy subjects, as well as in obese patients with or without T2DM (13). In the present study, the obesity risk rs17848939 G-allele was nominally significantly associated with the visceral to subcutaneous FASN mRNA expression ratio. Consistent with the previously described positive correlation of fat mRNA expression with measures of obesity, the rs17848939 variant also showed nominally significant association with BMI and % body fat (data not shown). These data suggest that genetic variation in FASN may at least partially explain variation in mRNA expression in visceral and subcutaneous adipose tissue. However, it needs to be noted that based on correlation data including mRNA expression together with genetic variation and a certain phenotype (such as BMI) one can not reveal the causative chain and so, to rule out that changes in mRNA expression simply reflect the state of obesity. As shown recently in a genome-wide study on mRNA expression in adipose tissue (23), the majority of the traits profiled (over 70% or 17.080 traits) showed significant correlation with BMI. Therefore, studies targeting functional consequences of FASN obesity risk variants on FASN transcription will be inevitable to clarify whether or not genetic variants can explain variation in mRNA expression.
In addition to above mentioned polymorphisms, we attempted to replicate previously reported correlations between the Val1483Ile substitution and obesity despite low MAF (4%) of this variant in Europeans of German origin. The variant was first identified and shown to be associated with percentage of body fat and substrate oxidation rates in Pima Indians (11). It has been hypothesised that the Val/Ile substitution, which is positioned in the FASN interdomain region (24) could alter this functionally essential site and thus potentially reduce FASN activity (11). Consequently, changes in FASN activity could affect substrate oxidation rates, which would result in the decrease in body fat observed in the Pima Indians (11). Further support for the protective effect of this variant against obesity came from studies in German children showing reduced BMI-SDS and improved lipid profile of boys carrying the Ile-variant (12). Findings in children cohorts suggested sex-specific effects of the polymorphism which was confirmed recently by independent studies in Spanish whites (25). Spanish men with the Ile-allele had lower BMI, WHR, fasting glucose, and blood pressure, and they showed increased insulin sensitivity (25). However, no association was found in women (25). Also in our study, in contrast to women, German adult men carrying the Ile-allele seem to be protected against obesity as suggested by the case–control study as well as by the association with lower BMI. Because Ile-carriers also have decreased adipose tissue FASN activity (25), it might be postulated that Val1483Ile is a functional variant in FASN. Sexual dimorphism in genetic associations in obesity, glucose, and lipid metabolism is not uncommon and has been reported for genes such as leptin (26), resistin (27), winged helix/forkhead transcription factor (28), peptide tyrosine tyrosine (29), Y2 receptor (29) and others. It could be hypothesised that the protective effects of the Val1483Ile are masked by hormonally controlled mechanisms in women.
It needs to be mentioned though that several other variants (with very low MAF) identified by sequencing and which predict amino acid substitutions were not examined in the present study. Therefore, we can not exclude that rare variants other than Val1483Ile might be affecting human obesity as well.
Children represent a valuable study population for identifying primary genetic determinants involved in susceptibility to complex polygenic diseases such as obesity. Unlike in adults their phenotype is less influenced by comorbidities and prolonged exposure to environmental factors. Interestingly, although we previously showed sex-specific association of the Val1483Ile variant with obesity in German children (12), no relationship with other SNPs associated with obesity in adults, was seen in our present study. Lack of association of FASN SNPs with childhood obesity indicates that the SNP effects most likely manifest in later stages of life and/or that these effects might be modified by gene-environment interactions. This aspect may need to be considered in association studies and may partially explain why genetic variation in FASN was not found to be associated with obesity in genome-wide association scans (GWAS). However, we are aware that effect sizes observed in the present study should have been picked up easily in well powered GWAS. Due to the relatively small sample size in our study, we see these effect sizes with caution. Also, we attempted to replicate the observed associations in our GWAS in a self-contained population of Sorbs from Germany (30). The GWAS was performed by using 500 K and 6.0 Affymetrix GeneChips. Unfortunately, no SNPs within the FASN locus were included on the chips, which made potential comparisons with our sequenced variants impossible. Similarly, the 1958 British Birth Cohort (BC58) publicly available data only provide three SNPs within the FASN (http:www.b58cgene.sgul.ac.ukindex.php). Thus the lack of associations with obesity in GWAS may partly be attributed to missing SNPs on the used GeneChip arrays. Further, similar MAF for SNPs rs62078748, rs2229422, rs17848939, rs2229425 in our control sample and HapMap populations might eventually support our association findings. Unfortunately, we were not able to compare our control sample with other populations in silico, since there are no publicly available data on allele frequencies for these SNPs at the moment.
We are aware that our findings will need to be confirmed in independent studies. However, it should be noted that the strongest association signal observed for rs2229422 in our study did withstand very conservative Bonferroni corrections for multiple testing, which would require P value of 0.006 for analyses of associations in the present case–control studies. In addition, we could replicate previously described protective effects of the Val1483Ile variant against obesity, which further supports validity of these findings.
Based on our data, we suggest that variation in the human FASN may play a role in the regulation of human body weight. Although these data support previous studies in rodents and replicate protective effects of the Val1483Ile against obesity, associations of common variants in FASN with obesity are novel and need to be replicated to confirm these findings and define their relative importance in the pathogenesis of human obesity.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http:www.nature.comoby
Acknowledgments
We thank all those who participated in the studies. We also thank Roy Tauscher for excellent technical assistance. This work was supported by grants from Deutsche Forschungsgemeinschaft, the Clinical Research Group “Atherobesity” KFO 152 (projects BL 833/1-1 to M.B., and Stu192/6-1 (M.S.), KO 3512/1-1 to A.K. and TP5 to W.K.), the Interdisciplinary Centre for Clinical Research Leipzig at the Faculty of Medicine of the University of Leipzig (Project N06 to P.K., D.S., B.E., K.D. and project B24 to M.B., B27 to M.S. and A.T.), from the German Diabetes Association (to Y.B., A.T., P.K., A.K.), from the Deutsche Hochdruckliga and by unrestricted grants from Merck Serono, Ipsen and Novo Nordisk (to W.K.).
Disclosure
The authors declared no conflict of interest.





