Genetic polymorphism of the type A human natriuretic peptide receptor (NPR-A) gene contributes to the interindividual variability in the BNP system
Abstract
Aims:
To analyse the contribution of recently described genetic polymorphisms in the human natriuretic peptide receptor (NPR-A) to the interindividual variability in the BNP system.
Methods and results:
We evaluated NT-proBNP in 402 subjects, including healthy controls (n=93), patients with acute coronary syndrome (n=194) and heart failure (n=115). Three polymorphic sites encoding six common haplotypes of the NPR-A receptor gene, including three haplotypes in the 5′ region (CT11, CT10 and CT6) and three haplotypes in the 3′ region (3-plus, 4-minus and 4-plus), were studied.
The frequency of the identified “4-minus” haplotype was higher in control subjects with high NT-proBNP (>75th percentile) levels as compared to those with low NT-proBNP levels (15.2% vs. 5.7%, p<0.05). In the control subjects, carriers of the “-plus/4-minus” genotype had about 2-fold higher median NT-proBNP levels than individuals with other genetic variants (142pg/ml (88–371pg/ml) vs. 71pg/ml (35–111pg/ml, p=0.011). In contrast, in patients with cardiovascular disorders no relation between NT-proBNP and the described polymorphisms was observed.
Conclusion:
The “4-minus” haplotype of the NPR-A receptor gene is associated with high NT-proBNP values and is a genetic determinant of the interindividual variability in the BNP system in healthy individuals but probably not in patients with cardiovascular disorders.
1. Introduction
B-type natriuretic peptide (BNP) and its N-terminal fragment (NT-proBNP) are neurohormones synthesized and secreted in equal proportions from the atrial and ventricular myocardium. The most important biological properties of BNP are peripheral vasodilatation and an increase in natriuresis/diuresis. Therefore, BNP is an important regulator of blood pressure and volume homeostasis [1].
Stimulus for BNP release is an increase in left ventricular wall stress [2], therefore plasma/serum levels are affected by the degree of myocardial stretch, damage, and ischaemia in the ventricle [3]. BNP and NT-proBNP, which can be measured in serum and plasma with commercially available assays, have been recently recognized as powerful diagnostic markers in several cardiovascular conditions. In patients with heart failure, increased concentrations of BNP and NT-proBNP are correlated with the haemodynamic status and clinical manifestation of the disease and accurately predict left ventricular ejection fraction as well as morbidity and mortality [4 5 6 7]. BNP and NT-proBNP are also known to provide valuable diagnostic and prognostic information in patients with acute coronary syndrome, valvular heart disease and pulmonary embolism and are used in these patients for risk stratification [8 9 10 11 12 13]. The diagnostic application of these markers in routine practice, however, is currently limited because of intra- and interindividual biological variations in the plasma/serum concentrations of the hormones [14] as well as insufficient knowledge about the factors responsible for this variability.
The inherited variations in the genes encoding a ligand receptor may be an important determinant in the variability of the ligand activity and influence its biological effects. The biological effects of BNP are mainly mediated by its binding to the natriuretic peptide receptor type A (NPR-A) leading to activation of guanylyl cyclase-A and an increase in cGMP. It has been demonstrated that the quantitative changes in the expression of the NPR-1 gene coding the expression of the NPR-A receptor have significant effects on blood pressure and cardiac mass in mouse models [15].
In humans, ten polymorphic sites forming three haplotypes in the 5′ and three haplotypes in the 3′ noncoding regions of the NPR-1 gene have been identified recently [16]. Transient expression analysis in cultured cells has revealed that one of the 5′ haplotypes, represented as the variant “CT6”, has 45% higher promoter activity than two other identified 5′ haplotypes (represented as the variants “CT10” and “CT11”). In the 3′UTR, two haplotypes (represented as the variants “3-plus” and “4-minus”) have been demonstrated to reduce the reporter gene function by 30%, as compared to the most common 3′-haplotype (represented as the variant “4-plus”). The genotype-phenotype relationship for these polymorphisms in vivo, however, has not yet been characterized.
It has been demonstrated that Japanese subjects genetically deficient in NPR-A receptors may exhibit higher levels of plasma BNP as compared to subjects with wild-type receptors [17]. Based on this observation, the aim of the present study was to examine whether the recently identified functional polymorphisms of the NPR-A receptor gene in the 5′ and 3′ region may also contribute to the reported interindividual variations in the activity of the BNP system. Therefore, we investigated the association between these common haplotypes of the gene and serum NT-proBNP levels in populations with normal and elevated NT-proBNP values.
2. Materials and methods
2.1. Study population
A total of 402 unrelated Caucasian individuals were recruited into the study after giving informed written consent according to the protocol approved by the local Ethics Committee of the Medical Faculty of the University of Frankfurt. There were three study groups; (1) patients with heart failure (HF) with a LVEF <50%, (2) patients with acute coronary syndrome and (3) control subjects in whom left ventricular dysfunction or relevant coronary artery stenosis had been excluded by echocardiography and angiography, respectively. ACS and HF patients were recruited from our coronary care unit and heart failure clinic, respectively. Control subjects were randomly chosen from patients admitted for diagnostic procedures. An acute coronary syndrome was defined as typical chest pain within the last 48 h with ischaemic changes in the ECG namely either ST segment elevation or ST segment depression in at least two contiguous leads or elevation of cardiac troponin T. Heart failure patients were recruited from our heart failure clinic during a regular visit.
2.2. Measurements of NT-proBNP and cGMP
In all patients blood samples were collected from an antecubital vein for determination of NT-proBNP levels. Samples were taken on admission, which was between 08.30 and 12.30 h for elective heart failure and control patients and anytime during 24 h for the ACS patients. The specimens were centrifuged (3000 g for 10 min) and frozen at −80 ° until analysis. Serum concentrations of NT-proBNP were measured using the electrochemiluminescence-immunoassay (Elecsys proBNP, Roche Diagnostics, Mannheim, Germany). The lower limit of detection was 5 pg/ml. The intra-assay coefficient of variation (CV) was 1.8% and 2.7% and the inter-assay CV was 2.3% and 3.2%. The concentration range tested was from 175 to 4962 pg/ml.
Additional EDTA blood was sampled from 40 randomly chosen individuals in the control group to determine plasma cGMP levels. The measurements were performed using a commercially available EIA using a polyclonal rabbit antibody (Assay Designs Inc, Ann Arbor, USA). The intra-assay CV was 5.2% (1.5 pmol/l), 4.0% (16.6 pmol/l) and 7.6% (481 pmol/l). The inter-assay CV was 13.7% (1.8 pmol/l), 3.5% (16.9 pmol/l) and 5% (359 pmol/l).
2.3. Haplotyping of the NPR-1 gene
Genomic DNA was isolated in all patients using a High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany). The NPR-1 gene was haplotyped using the novel non-gel-based high throughput sequencing-by-synthesis technique, called pyrosequencing [18,19].
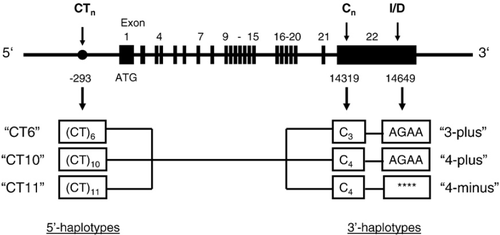
Three polymorphic sites in the NPR-1 gene sequence used for the haplotyping were adapted from Knowels et al. [16]. In particular, CTn dinucleotide repeats (n=6, 10, or 11) at nucleotide −293 in the promoter region (“CTn polymorphism”) was used to determine 5′ haplotype. A length polymorphism of 3C or 4C at nucleotide 14,319 in the 3′UTR (“Cn polymorphism”) and a 4-bp insertion/deletion of AGAA at nucleotide 14,649 (“I/D polymorphism”) in the 3′UTR were selected to determine 3′ haplotypes (Fig. 1). Each polymorphism was amplified from the genomic DNA (60 ng) by separate PCR in the presence of 0.5 μM of forward and reverse primers and 25 μl of HotStarTaq MasterMix Kit (Qiagen, Hilden, Germany) in a total volume of 50 μl, generating 107-bp (CTn polymorphism), 146-bp (Cn polymorphism) and 127-bp (I/D polymorphism) fragments. One of the primers in the respective set was biotinylated at the 5′-end to allow immobilization of the PCR product in the subsequent sequencing reaction. The PCR reaction was started by initial denaturation at 95 °C for 15 min. Amplification was performed for 50 cycles including denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min and extension at 72 °C for 1 min.
The biotinylated PCR product (25 μl) was then immobilized to streptavidin-coated Sepharose beads (Amerhsam Biosciences Europe GmbH, Freiburg, Germany) in the presence of the binding buffer (10 mM Tris-HCl, 2 M NaCl, 1 mM EDTA, 0.1% Tween 20, pH 7.6). The template preparation was performed using Vacuum Prep Tool (Biotage, Uppsala, Sweden). Single-stranded DNA was obtained by short incubation of the immobilized PCR product in 70% Ethanol and alkali treatment with 0.2 M NaOH, followed by washing with the buffer (10 mM Tris-Acetate, pH 7.6) to remove the leftover from the PCR reaction. The single-stranded template was then resuspended in the sequencing mixture (40 μl) containing annealing buffer (20 mM Tris-Acetate, 2 mM Mg-Acetate, pH 7.6) and 0.4 μM of the respective sequencing primer in a 96-well PSQ 96 plate. The sequencing primer was allowed to anneal on a heat plate set at 80 °C for 2 min and then cooled to room temperature. The sequencing reaction was performed at 25 °C on the automated pyrosequencer instrument (Biotage, Uppsala, Sweden) using PSQ 96 SNP Reagent Kit (Biotage, Uppsala, Sweden) according to the manufacturer's instructions.
The detection of the CTn polymorphism in the 5′ haplotype was performed using the PCR primers CTnfor (5′CGCCCTCTCTCATCCTTCTTC3′) and CTnrev (5′ biotin GAGGACCCGAGTGTGAACAAC3′) as well as the sequencing primer CTnseq (5′CACTCGCACCCTTT3′). The detection of the Cn polymorphism in the 3′ haplotype was performed using the PCR primers Cnfor (5′ biotin CTGTGGATTCCTGATCCCCTC3′) and Cnrev (5′GCCCTCTCTTCACCTAGTCTC3′). The sequencing primer was Cnseq (5′GGCACAGGGTAGGG3′). For the detection of the I/D polymorphism in the 3′ haplotype the amplification primers I/Dfor (5′ biotin CTGTGGATTCCTGATCCCCTC3′) and I/Drev (5′GCCCTCTCTTCACCTAGTCTC3′) were used. The sequencing reaction was performed using the primer I/Dseq (5′TTCAGGTGA-CTCT3′).
2.4. Statistical methods
Serum concentrations of NT-proBNP are presented as median and interquartile range (IQR). Other variables are given as mean and standard deviation (SD). The comparisons of NT-proBNP levels were performed using Kruskal-Wallis test and Mann-Whitney test. Other parameters were compared using the Student's t-test or ANOVA. For multiple comparisons Bonferroni test and the Dunnetts-T3 test have been applied. The correlation between NT-proBNP and cGMP at baseline was tested using the Spearman correlation coefficient. The comparison of categorical variables was performed using the χ2-test. To visualize the relationship between NT-proBNP levels and the genetic variants of the NPR-A receptor gene, patients were subdivided by the 75th percentiles of their BNP values. The association between the dichotomised NT-proBNP values and the genetic variants of the NPR-A receptor gene was evaluated using Fischer's exact test. To analyse whether the “4-minus” NPR-A polymorphisms is an independent determinate for elevated NT-proBNP, we performed stepwise multivariate logistic regression analyses using age, gender, body mass index, left ventricular end diastolic pressure and the prevalence of “4-minus” NPR-A polymorphism as explanatory variables. The statistical analysis was performed using SPSS 10.0 software (SPSS Inc., Chicago, USA). A p-value (two-tailed) <0.05 was considered statistically significant.
3. Results
3.1. Patient characteristics and NT-proBNP values
The study population consisted of 402 subjects, 194 patients with acute coronary syndrome, 115 patients with HF and 93 controls. The baseline characteristics of the included individuals are shown in Table 1. There was a male predominance in the ACS and HF groups as compared to the control group. ACS patients were older (66±12 years) than heart failure patients (62±10 years; p=0.016) and controls (61±10 years; p=0.002). Systolic blood pressure was higher in the controls (158±24 mm Hg), as compared to ACS patients (138±30 mm Hg; p<0.001) and HF patients (126±19 mm Hg; p<0.001). Heart rate was comparable in controls (75±12/min) and ACS patients (78±16 /min; p=0.434) but was significantly lower in heart failure patients (69±15 /min; p=0.012).
| Controls | ACS | Heart failure | p-value | |
|---|---|---|---|---|
| n | 93 | 194 | 115 | |
| Male/female | 34/59 | 125/69 | 98/17 | <0.001 |
| Male (%) | 36.6% | 64.4% | 85.2% | <0.001 |
| Age (years) | 61±10 | 66±12 | 62±11 | 0.001 |
| Age>65 years (%) | 39% | 58% | 47% | 0.008 |
| Body mass index (kg/m2) | 28.3±4.9 | 27.4±4.5 | 28.7±15.1 | 0.458 |
| Systolic blood pressure (mm Hg) | 158±24 | 137±30 | 126±19 | <0.001 |
| Heart rate (/min) | 75±12 | 78±16 | 69±15 | <0.001 |
| Ejection fraction (%) | >50% | Nd | 27±16 | <0.001 |
| NT-proBNP (pg/ml) | 75 (41–131) | 841 (195–2354) | 831 (319–2336) | <0.002 |
- a Values are expressed as absolute or relative numbers, as mean±standard deviation. NT-proBNP is expressed as median with the interquartile range in parentheses.
Median (25th to 75th percentile) NT-proBNP levels were within the normal range in the control patients. As expected, NT-proBNP concentrations were markedly increased in the ACS and HF patients (Table 1). NT-proBNP levels were linked to sex, with females having higher concentrations than males in the control group — 92 pg/ml (57–182 pg/ml) vs. 54 pg/ml (21–91 pg/ml); p=0.001 and in the ACS group — 1898 pg/ml (245–3434 pg/ml) vs. 568 pg/ml (180–1831 pg/ml); p=0.001. No sex related differences were observed in the HF patients: 831 pg/ml (319–2439 pg/ml) vs. 733 pg/ml (271–1690 pg/ml), p=0.599, this may have been due to the significantly higher LVEF values in females compared to males in this group (36±15% vs. 26±15%; p=0.022). Patients over 65 years old had higher values of NT-proBNP than those younger than 65 years, both in the HF group (1374 pg/ml (480–3045 pg/ml) vs. 609 pg/ml (244–1317 pg/ml); p=0.002) and in the ACS group (1809 pg/ml (367–3031 pg/ml) vs. 362 pg/ml (99–966 pg/ml); p<0.001). In the control group, there was a strong trend towards higher NT-proBNP values in the patients older than 65 years (71 pg/ml (29–102 pg/ml) vs. 95 pg/ml (49–161 pg/ml); p=0.062).
3.2. The frequency of NPR-A haplotypes/ genotypes
Six common haplotypes, including three haplotypes in the 5′ region and three haplotypes in the 3′ region of the NPR-A receptor gene, were identified in a total of 402 DNA samples. The 5′ haplotypes represented by the variants, “CT6”, “CT10” and “CT11”, were detected with approximate overall frequencies of 0.1%, 48.2% and 51.7%, respectively. The 3′ haplotypes were distributed as follows: “3-plus” 46.9%, “4-plus” 46.8% and “4-minus” 6.3%. The distribution of the genotypes was in agreement with the Hardy-Weinberg equation, where the expected genotype frequencies were within the 95% confidence interval of those found (data not shown). The frequencies of the studied haplotypes were comparable in all three study groups. In particular, the frequencies of the 5′ genotypes in the control group, ASC patients and HF patients were 19%, 20% and 22% for CT10/CT10, respectively, 56%, 59% and 48% for CT10/CT11, respectively, 24%, 21% and 30% for CT11/CT11, respectively and 1%, 0% and 0% for CT6/CT11, respectively. The frequencies of the 3′ genotype in the control group, ASC patients and HF patients were 23%, 19% and 28% for “3-plus/3-plus”, respectively, 42%, 43% and 45% for “3-plus/4-plus”, respectively, 19%, 25% and 18% for “4-plus/4-plus”, respectively, 8%, 8% and 3% for “3-plus/4-minus”, respectively, and 9%, 6% and 5% for “4-plus/4-minus” genotypes, respectively. There were no sex related differences observed in the distribution of the established genotypes (χ2=3.31, p=0.507 for the 3′ genotypes and χ2=0.925, p=0.630 for the 5′ genotypes). Moreover, there were no differences in the frequency of the “4-minus” haplotype between males and females (χ2=1.66, p=0.197) and between patients >65 years old and those <65 years old (χ2=1.33, p=0.249).
3.3. Association between the genetic variants of the NPR-A receptor gene and NT-proBNP levels
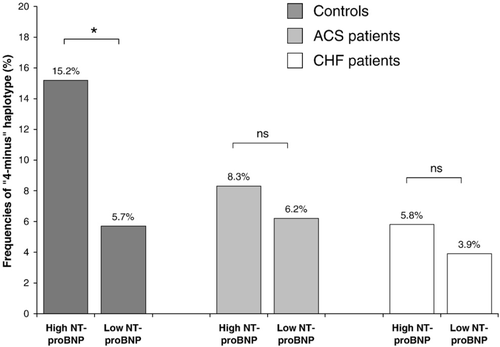
The 5′ haplotypes had no effect on NT-proBNP levels. In the 3′ region, the “4-minus” haplotype was associated with increased NT-proBNP levels (by quartile) in all groups; however, this association was statistically significant only in healthy controls. The frequencies of the“4-minus” haplotype among the individuals with high NT-proBNP levels (>75th percentile) as compared to those with NT-proBNP values <75th percentile were as follows: 15.2% vs. 5.7% in controls (p=0.048), 8.3% vs. 6.2% in patients with ACS (p=0.467) and 5.8% vs. 3.9% in patients with HF (p=0.692) (Fig. 2). In the stepwise multivariate logistic regression analysis using age, sex, body mass index, left ventricular end diastolic pressure (LVEDP) and the prevalence of the “4-minus” NPR-A polymorphism as explanatory variables for elevated NT-proBNP concentrations in healthy controls, the “4-minus” haplotype was an independent determinant for NT-proBNP elevation above the 75th percentile (p=0.018) together with sex (p=0.032).
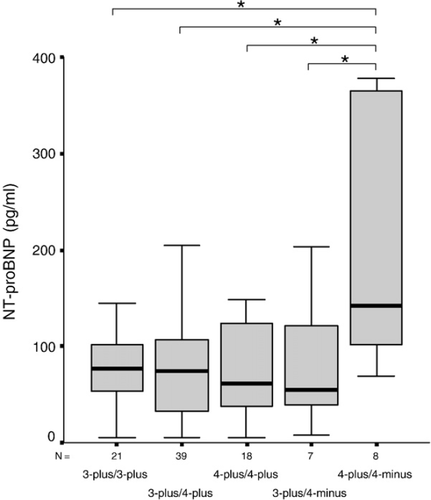
A marked difference in NT-proBNP level was found in the control subjects with “4-plus/4-minus” genotype, its carriers had about 2-fold higher median serum NT-proBNP levels compared to the individuals with other genotypes (142 pg/ml (88–371 pg/ml) vs. 71 pg/ml (35–111 pg/ml), p=0.011) (Fig. 3). When NT-proBNP levels were dichotomised by the 75th percentile (131 pg/ml), the frequency of the “4-plus/4-minus” genotype was more than 5-fold higher in individuals with NT-proBNP >131 pg/ml than those with NT-proBNP <131 pg/ml (p=0.021). No associations between any particular 3′ or 5′ genotypes and NT-proBNP levels were observed in ACS or HF patients (Tables 2 and 3).
| Controls | ACS | Heart failure | |
|---|---|---|---|
| 5′ genotypes | |||
| CT10/CT10 | 86 (57–183) | 568 (238–2337) | 573 (246–1458) |
| CT10/CT11 | 64 (39–106) | 1084 (195–2354) | 762 (249–2251) |
| CT11/CT11 | 86 (26–178) | 573 (162–2566) | 1314 (396–3890) |
| CT6/CT11 (n=1) | 20 | ||
| 3′ genotypes | |||
| 3-plus/3-plus | 77 (41–109) | 1396 (158–2617) | 997 (247–1851) |
| 3-plus/4-plus | 75 (28–107) | 803 (162–2026) | 1080 (397–2420) |
| 4-plus/4-plus | 62 (36–130) | 654 (247–2350) | 426 (109–1394) |
| 3-plus/4-minus | 54 (31–185) | 1637 (364–5319) | 388 (255–700) |
| 4-plus/4-minus | 142 (88–371) | 518 (302–2395) | 1950 (772–3560) |
- a Values are expressed as median with interquartile range in parentheses.
| NT-proBNP (pg/ml) | no 4-plus/4-minus | 4-plus/4-minus | p-value |
|---|---|---|---|
| Controls | |||
| ≤131 (75th perc.) | 67 (96%) | 3 (4%) | p=0.021 |
| >131 | 18 (78%) | 5 (22%) | |
| ≤288 (90th perc.) | 79 (94%) | 5 (6%) | p=0.028 |
| >288 | 6 (67%) | 3 (33%) | |
| ACS | |||
| ≤2364 (75th perc.) | 138 (95%) | 8 (5%) | p=1.000 |
| >2364 | 45 (94%) | 3 (6%) | |
| ≤4958 (90th perc.) | 164 (94%) | 11 (6%) | p=0.605 |
| >4958 | 19 (100%) | 0 (0%) | |
| Heart failure | |||
| ≤2369 (75th perc.) | 86 (97%) | 3 (3%) | p=0.128 |
| >2369 | 23 (88%) | 3 (12%) | |
| ≤5961 (90th perc.) | 104 (94%) | 6 (6%) | p=1.000 |
| >5961 | 11 (100%) | 0 (0%) | |
- a Values are expressed as absolute numbers and percentage in parenthesis. p-value refers to Fischer's exact test.
3.4. cGMP and NPR-A genotypes
In 40 randomly chosen individuals from the control group, the median cGMP value was 0.049 pmol/ml with an interquartile range of 0.040 to 0.058 pmol/ml. There was no association between cGMP values and the NPR-A receptor gene haplotypes or sex. Moreover, we did not find a correlation between NT-proBNP and systemic cGMP levels (r=0.084, p=0.608).
4. Discussion
Human type A natriuretic peptide receptor (NPR-A) mediates the biological functions of the natriuretic peptides ANP and BNP and is a candidate gene for affecting the incidence and severity of several cardiac disorders. Three common haplotypes in the 5′ (“CT6”, “CT10” and “CT11”) and 3′ (“3-plus”, “4-plus” and “4-minus”) regions of the receptor gene have been identified recently [16]. Three of these, including “CT6”, “3-plus” and “4-minus”, have been shown to have quantitative effects on the expression of the receptor in vitro. In the present study, we analysed the functional impact of these polymorphisms in vivo by studying their relationship to serum NT-proBNP levels in controls and in patients with ACS and HF.
The major finding of the study is that the “4-minus” haplotype, identified in the 3′-untranslated region of the NPR-A receptor gene, is associated with significantly higher NT-proBNP levels as compared to other genetic variants. The “4-minus” haplotype, which is detected with an overall frequency of 6.3%, is constituted by 4C repeats at nucleotide 14,319 and a 4-bp deletion of AGAA at nucleotide 14,649. The transient expression analysis in cultured cells of reporter plasmids with the 3′ regions has shown that this genetic variant reduces the gene expression of the NPR-A receptor gene by 30% as compared to the “4-plus” haplotype [16]. The mechanism responsible for this effect is supposed to be mRNA instability leading to the decreased translation of the gene.
In our study, the individuals with the genetically deficient NPR-A receptor due to the presence of the “4-minus” haplotype exhibited much higher NT-proBNP levels than those without this defect. We suppose that this observed association is the result of negative feedback regulation, where the diminished function of the BNP system caused by the genetic defect in its receptor consequently leads to the up-regulation of the BNP gene expression and a secondary release of BNP in order to compensate for the deficiency. A similar autoregulation mechanism has been described for atrial natriuretic peptide (ANP), which is from the same protein family with a similar structure and function and is another ligand for the NPR-A receptor [20]. In particular, it has been demonstrated that NPR-A deleted transgenic mice show more than two-fold increased plasma ANP levels as compared with wild types [21]. In humans, functional deletion of the NPR-A receptor gene has been shown to be associated with significantly higher BNP levels compared to wild type [17].
The observed relationship between “4-minus” haplotype and high NT-proBNP levels was statistically significant only in the control group, and we did not observe a similar relationship in either of the patient groups. It is likely that this lack of significance is due to the influence of confounding factors related to the cardiovascular disorders (i.e. degree of ventricular dysfunction, magnitude of the myocardial ischaemia, creatinine values etc.), which are known to affect BNP levels and which therefore may have biased the established genotype-phenotype relationship. In cardiovascular patients, BNP and NT-proBNP are often already maximally up-regulated as a consequence of haemodynamic alterations and myocardial ischaemia. Therefore the impact of the NPR-A receptor defect on the BNP regulation in such cases may be difficult to detect. Our data suggest that the described polymorphism has no effect on interindividual variations in BNP or NT-proBNP values under pathological conditions.
In addition to the measurement of NT-proBNP levels, we also evaluated plasma concentrations of cGMP to assess whether this marker can be used as an intermediate phenotype for NPR-A receptor gene polymorphisms. cGMP is the intracellular second messenger of the NPR-A and cGMP levels have been shown to correlate with endogenous natriuretic peptides and exogenous BNP administration [22,23]. We could not find any relationship between systemic cGMP values, NT-proBNP levels and NPR-A receptor gene polymorphisms. There may be several reasons for the observed finding. Firstly, cGMP is a second messenger for several other hormones, including CNP and NO, and its regulation is complex. Secondly, the intracellular concentrations of cGMP may significantly differ from their circulating concentrations. Therefore, plasma cGMP is not an appropriate marker to assess the function of the NPR-A polymorphisms.
The established relationship between the “4-minus” haplotype and high NT-proBNP levels provides new insights into the diagnostic and therapeutic applications of BNP. Firstly, the “4-minus” haplotype appears to be an important determinant contributing to the interindividual variations of BNP levels. An understanding of such determinants is essential for interpreting test results, particularly when these tests are obtained in the non-acute setting, where marked elevations are uncommon. The impact of other factors (i.e. sex, age) on NT-proBNP and BNP levels have been extensively described [24 25 26 27]. In keeping with these findings, we also found higher NT-proBNP levels in females from the control and ACS groups. The absence of a sex related difference in HF patients was probably due to the higher LVEF values in the females compared to the males in this group. Importantly, the “4-minus” polymorphism was not related to sex or age, so that the demographic differences in the three study groups as a result of the epidemiological peculiarities of the underlying heart diseases could not bias the results of the study. Therefore, the “4-minus” haplotype is an independent predictor of high NT-proBNP values.
Another important area where the established genotype-phenotype relationship may be of clinical relevance is pharmacotherapy with recombinant natriuretic peptides such as nesiritide in patients with acutely decompensated heart failure. Future studies should evaluate whether a deficient “4-minus” haplotype is more frequent in non-responders to this treatment and, therefore, whether this may be responsible for the therapeutic failure in some patients. Finally, the “4-minus” polymorphism of the BNP receptors may have important biological consequences and its effect on the incidence, severity and prognosis of several cardiac disorders, including arterial hypertension, acute coronary syndrome and heart failure, should be evaluated.
5. Limitations
There are some limitations to our study which need to be addressed. The sample size was relatively small. However, since these patients comprised an unselected cohort recruited from daily routine, the achieved results are representative. Concomitant medication was not assessed systematically, therefore we cannot be certain that drug use was similar between patients with the various polymorphisms. However, this would have occurred by chance and a systematic bias of the results is unlikely. We were not able to analyse all polymorphisms which have been described recently. Thus it is possible that other polymorphisms could have influenced NT-proBNP serum levels to a different extent as well.
6. Conclusion
This is the first study to show that the “4-minus” haplotype of the NPR-A receptor gene, which is related to the decreased expression of the receptor in vitro, is associated with high NT-proBNP values in vivo. The established genotype-phenotype relationship is an important determinant of the interindividual variability in the BNP system in apparently healthy individuals and provides new insights into the diagnostic and therapeutic applications of BNP testing. However, the described polymorphism has no effect on interindividual variations in BNP or NT-proBNP values in patients with cardiovascular disorders.
7 Acknowledgements
The study was supported by the DFG, Excellence Cluster Cardiopulmonary System.
We would like to thank Martin Deile, Henning Brück, Matthias Lintz, Claudiu Oltenau and Philip Kahler for the excellent and enthusiastic work they did in the clinical chemistry and biomolecular laboratories. Furthermore, we are indebted to Prof. Dragic Bankovic from the mathematical faculty of Kragujevac, Serbia & Montenegro for the statistical review and advice. This study has been supported by Roche diagnostics that has granted NT-proBNP assays without charge. The study has been performed without external financial support.




