Cloning and functional analysis of the orotidine-5′-phosphate decarboxylase gene (PbrURA3) of the pathogenic fungus Paracoccidioides brasiliensis
Abstract
A genomic clone encoding the Paracoccidioides brasiliensis orotidine monophosphate decarboxylase gene (PbrURA3) was isolated by screening a subgenomic plasmid DNA library of this fungus, using a PCR amplification product of the gene as a probe. Sequence analysis revealed that the gene contains an open reading frame of 855 bp with a single intron (162 bp), and encodes a putative 285 amino acids polypeptide of estimated molecular weight 31.1 kDa and isoelectric point 6.5. The deduced amino acid sequence predicted a 73.4% identity with orotidine monophosphate decarboxylase of Aspergillus nidulans. Functionality of the gene was demonstrated by transformation into a Saccharomyces cerevisiae ura3 null mutant. The PbrURA3 gene sequence has been submitted to GenBank database under Accession No. AJ133782. Copyright © 2005 John Wiley & Sons, Ltd.
Introduction
Orotidine-5′-monophosphate decarboxylase is an enzyme responsible for the conversion of orotidine-5′-phosphate (OMP) to uridine-5′-phosphate (UMP), the final step in the pyrimidine biosynthetic pathway. It is encoded by URA3, a gene that was initially used to derive a construct (the ura-blaster cassette; Alani et al., 1987) for the disruption of Saccharomyces cerevisiae genes.
In order to study the molecular mechanisms underlying pathogenicity and other fungal biological processes, the disruption technique became particularly attractive in the study of Candida albicans (Fonzi and Irwin, 1993; Goshorn et al., 1992; Lay et al., 1998; Bain et al., 2001; Sundstrom et al., 2002). Also, in non-C. albicans species the disruption of alleles in diploid strains and multiple gene disruption has been reported (Sakai et al., 1991; Zhou et al., 1994; Rodríguez et al., 1998). The pathogenic dimorphic fungus Paracoccidioides brasiliensis is the causative agent of paracoccidioidomycosis, one of the most frequent systemic mycoses in the rural population of Central and South America. This fungus can grow as hyphae at 20–23 °C and change to yeast if a thermal stimulus at 37 °C occurs (San-Blas and Niño-Vega, 2001, 2004; San-Blas et al., 2002). Genomic studies (Reinoso et al., in press) and partial transcriptome analysis of P. brasiliensis (Felipe et al., 2003; Goldman et al., 2003) identified genes involved in cellular metabolism, information storage and cellular processes. Some of them were differentially expressed during the dimorphic transition, others were potentially involved in drug resistance and virulence. Additionally, P. brasiliensis genes related to dimorphism have been studied (for review, see San-Blas and Niño-Vega, 2004).
Functional studies on these genes by single and multiple gene disruptions could help to establish their real involvement in dimorphism and pathogenicity, since the development of efficient transformation systems is essential to characterize such genes and their contribution to the pathogenic process in this medically important fungus. A transformation system have recently been reported for P. brasiliensis (Leal et al., 2004). The present paper describes the cloning and complementation in yeast of the P. brasiliensis URA3 gene (PbrURA3) as a first step in the construction of a hisG_PbrURA3_hisG cassette and its use for gene disruptions in P. brasiliensis.
Materials and methods
Strains, media and growth conditions
P. brasiliensis strain IVIC Pb73 (ATCC 32 071) was grown in PYG (0.5% w/v bactopeptone, 0.5% w/v yeast extract, 1.5% w/v glucose) liquid medium. Cells were grown at either 23 °C (M form) or 37 °C (Y form) with shaking at 100 r.p.m. Escherichia coli XL1-Blue (Stratagene, Cambridge, UK), used for propagation of plasmids and cloning experiments, was grown in Luria–Bertani (LB) medium supplemented with 100 µg/ml ampicillin (Sambrook et al., 1989).
Nucleic acids isolation
Genomic DNA was isolated as previously described (Calcagno et al., 1998). Total RNA was prepared from ground cells using TRIzol (Life Technologies, Rockville, USA). Plasmid DNA extraction was done by the Qiagen (Chatsworth, USA) midi kit.
Isolation and sequencing of the PbrURA3 gene
To amplify a fragment of the P. brasiliensis URA3 gene, primers were designed on the basis of conserved Ura3 amino acid sequences from other fungal species (Figure 1). The following oligonucleotides were used: 5′-GTSYGGTACTGYTGMCCRWG-3′ and 5′-ATCTTCGARGAYCGCAARTTC-3′. The amplification reaction was performed with Taq polymerase (Promega, Madison, USA). Conditions for PCR were: 1 cycle at 95 °C for 4 min; 30 cycles at 95 °C for 1 min, 50 °C for 1 min, 72 °C for 2 min; 1 cycle 72 °C for 5 min. The PCR product was subcloned into the pGEM-T vector (Promega).
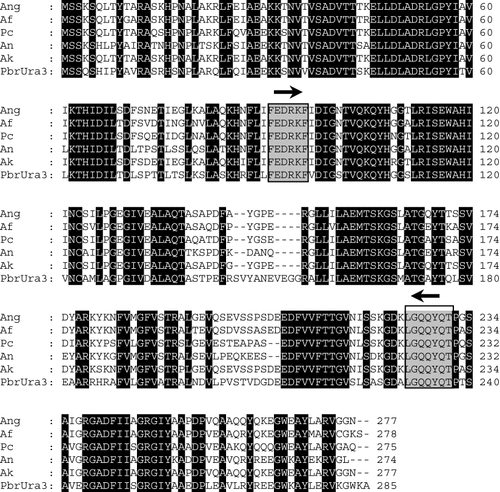
Multiple alignment of amino acid sequences from PbrURA3 and other highly homologous fungal orotidine-5′-phosphate decarboxylase proteins. Black boxes, areas of complete identity; grey boxes, conserved regions on which primers for partial amplification of PbrURA3 were designed; arrows over grey boxes, direction of primers. Ang, Aspergillus niger (Accession No. X96 734); Af, Aspergillus fumigatus (Accession No. Y11 303); Pc, Penicillium chrysogenum (Accession No. X08 037); An, Aspergillus nidulans (Accession No. M19132); Ak, Aspergillus kawachii (Accession No. AB064659); Pbr, Paracoccidioides brasiliensis (Accession No. AJ133782)
DNA (5–10 µg) was digested with several restriction enzymes. Digested DNA was subjected to 0.8% agarose gel electrophoresis in TBE buffer and blotted onto Hybond-N+ nylon membrane (Amersham, UK) according to Sambrook et al. (1989). Southern hybridization was performed with the amplified and cloned PbrURA3 radiolabelled fragment as a probe.
To clone the PbrURA3 gene, a genomic EcoRI library was constructed following the Southern analysis. Total DNA (80–100 µg) was digested with EcoRI and subjected to electrophoresis. A DNA fraction around 4.5 kb was extracted from the gel and ligated into the EcoRI site of pBluescript SK vector (Stratagene). Resulting transformants were collected in six pools, the recombinant plasmids were EcoRI-digested and analysed by Southern hybridization and one of the positive EcoRI libraries was screened by colony hybridization, using the labelled PCR fragment as probe (Sambrook et al., 1989). A positive clone was chosen for sequencing and further analysis.
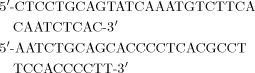
Cloning of the PbrURA3 into a S. cerevisiae expression vector
An 855 bp PCR product, which included the full-length ORF of the PbrURA3 gene, was synthesized according to the SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen Life Technologies, Carlsbad, CA, USA), using as gene-specific primers URA-EX1 (5′-ACA GAA TTC AAC CCC TCC ATC CAT-3′) and URA-EX2 (5′-TGG TCT AGA ACA CAG AGC ACC CCT-3′). The sense primer URA-EX1 contained the putative start codon and an engineered EcoRI restriction site (underlined); the antisense primer URA-EX2 contained the putative stop codon and an engineered XbaI restriction site (underlined). The 855 bp fragment was first cloned into TOPO 2.1 using the TOPO TA cloning kit (Invitrogen). In order to confirm that no mistakes were introduced by the enzyme, the product was subjected to automated sequencing. The resultant plasmid was digested with EcoRI and XbaI and subcloned into the galactose-inducible overexpression vector pYES6/CT (Invitrogen), which was similarly digested. Insertion of the gene in-frame with the C-terminal peptide was confirmed by automated sequencing.
Transformation of a S. cerevisiae null mutant with the PbrURA3 gene
The plasmid construct pYES6/CT–PbrURA3 was used to transform a S. cerevisiae ura3 null mutant (INVSc1 his3 leu2 trp1 ura3) using the Sc. EasyComp™ transformation kit (Invitrogen). A negative control was obtained by transformation of S. cerevisiae INVSc1 with pYES6/CT. Transformants were recovered and maintained on YPD agar plates (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) containing blasticidin (25 µg/ml) required for selection. Total DNA from four clones transformed with pYES6/CT and four clones transformed with pYES6/CT-PbrURA3, were extracted as described by Akada et al. (2000) and used for screening by PCR with primers URA-EX1 and URA-EX2. For further characterization, only one of the suspected Ura3+ transformants and one of the possible Ura3− transformants were chosen. Selected transformants were plate-replicated in synthetic medium SC (0.67% yeast nitrogen base plus amino acids, 2% dextrose, 2% agar), supplemented with blasticidin (10 µg/ml) and grown at 30 °C for 72 h. In order to test the complementation of the ura3 mutation, cells from each transformant were spread on minimal induction medium SC with or without 5-fluoroorotic acid (0.67% yeast nitrogen base containing histidine, leucine, tryptophan and uracil, 1% raffinose, 2% galactose, 2% agar), supplemented with blasticidin (10 µg/ml). 5-FOA was added at 1 mg/ml, as described by Boeke et al. (1984). Transformant cells were also grown in minimal induction medium SC without uracil.
Results and discussion
Characteristics of the P. brasiliensis URA3 gene
The PbrURA3 gene is made of 1017 bp, with an open reading frame (ORF) of 855 bp, interrupted by one intron (GenBank Accession No. AJ133782). RT-PCR confirmed the presence of the intron at positions +158 to +319 bp (162 bp), flanked by the conserved 5′-GT, 3′-AG and the characteristic splicing signal (lariat sequence) observed in fungal introns (Ballance, 1986). Putative TATA elements in the 5′-untranslated region of the PbrURA3 gene were found 592, 450 and 120 bp upstream from the proposed initiation codon. Possible CAAT boxes were located at 510, 265 and 157 bp upstream from the start codon. The sequences CACACA (−424) and CAACAA (−600 and −353) in PbrURA3 correspond to hexanucleotides present near the start codon in highly expressed genes (Davidow et al., 1987).
Analysis of the deduced amino acid sequence of PbrURA3
The identified ORF encodes for a putative 285 amino acids polypeptide, with a predicted molecular weight of 31.1 kDa and an isoelectric point of 6.5.
Sequence alignment with other Ura3 proteins showed high sequence identity between P. brasiliensis Ura3 and A. nidulans Ura3 (73.4%), followed by Penicillium chrysogenum (68.4%), A. niger (67.9%), A. fumigatus and A. kawachii Ura3 (67%) (Figure 1). Lower homologies were found for S. cerevisiae (47.6%), C. albicans (44.4%), and Schizosaccharomyces pombe Ura3 (42%).
Complementation of a S. cerevisiae ura3 null mutant strain by PbrURA3
The 855 bp cDNA, encoding the full-length Ura3 protein, was subcloned into the yeast expression vector pYES6/CT (Invitrogen), which contains a GAL1 promoter for inducible expression in yeast by galactose and repression by glucose. This construct was used to transform a S. cerevisiae ura3 null mutant. As a control, the mutant strain was transformed with the pYES6/CT plasmid alone. DNA from both transformants was isolated by a rapid method and used directly for PCR amplification. No signal was observed for the control Ura− transformants, when compared to the 855 bp PCR amplified fragment generated by the transformed Ura+ clones. To confirm the functionality of the PbrURA3 gene, transformants were further identified by their ability to grow on medium with or without uracil or 5-FOA, the latter being toxic when the URA3 gene is expressed. Both the pYES6/CT-transformed S. cerevisiae INVSc1 and the pYES6/CT + PbrURA3-transformed S. cerevisiae INVSc1 were able to grow in synthetic minimal medium plus uracil (Figure 2A). However, after 11 days of incubation at 30 °C, growth in the uracil-devoid medium was observed for the mutant transformed with pYES6/CT + PbrURA3, in opposition to the pYES6/CT-transformed S. cerevisiae INVSc1 that was unable to grow (Figure 2B). In contrast, when the minimal medium was supplemented with 5-FOA and uracil, the opposite result was obtained (Figure 2C). These results suggest that the putative P. brasiliensis URA3 gene complements the ura3 mutation in S. cerevisiae. The construction of an ura-blaster cassette using the PbrURA3 gene could facilitate the multiple disruption of P. brasiliensis genes and study their role in the related processes of pathogenicity and dimorphism.
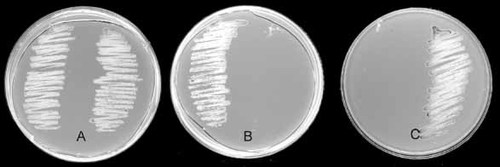
Complementation of a S. cerevisiae ura3 null mutant with the PbrURA3 gene. Growth in synthetic minimal medium (A) plus uracil, (B) minus uracil and (C) plus uracil and 5-FOA. Left side of each plate, S. cerevisiae INVSc1 strain transformed with pYES6/CT + PbrURA3; right side, S. cerevisiae INVSc1 strain transformed with pYES6/CT
Acknowledgements
C. Reinoso was a doctoral fellow supported by the Mycology Network, Project type B3, ALFA ProgramEuropean Union, No. ALR/B73011/94.05-5.0118.9 and CISET. The authors are grateful to Professor Germán Larriba (Universidad de Extremadura, Badajoz, Spain) for suggestions to improve this manuscript.




