LncRNA FOXP4-AS promotes the progression of non-small cell lung cancer by regulating the miR-3184-5p/EIF5A axis
Abstract
Long non coding RNA FOXP4-AS1 exerted crucial functions in various human cancers, while its role in non-small cell lung cancer (NSCLC) remains unclear. A total of 30 pairs of NSCLC tissues and matched adjacent normal tissues were used to evaluate the expression of FOXP4-AS1 and miR-3184-5p. Cell proliferation was assessed by CCK-8 assay and colony formation assay. Cell apoptosis was measured by flow cytometry. Bioinformatic analysis and luciferase reporter assay were performed to determine the regulatory relationship among FOXP4-AS1, miR-3184-5p and EIF5A. The xenograft tumor model was constructed to confirm the function of FOXP4-AS1 in NSCLC progression. The results showed that FOXP4-AS1 was upregulated and miR-3184-5p was downregulated in NSCLC tissues and cell lines. Downregulation of FOXP4-AS1 significantly reduced cell proliferation and induced apoptosis of NSCLC cells in vitro. FOXP4-AS1 could regulated the expression of EIF5A by binding to miR-3184-5p. Rescue experiments showed that downregulation of miR-3184-5p or overexpression of EIF5A obviously attenuated the inhibitory effects of si-FOXP4-AS1 on cell proliferation, as well as the stimulating effects on cell apoptosis. Moreover, knockdown of FOXP4-AS1 could efficiently inhibited tumor development of NSCLC in vivo. Downregulation of FOXP4-AS1 attenuated the progression of NSCLC by regulating miR-3184-5p and EIF5A.
1 INTRODUCTION
Lung cancer is the major leading cause of cancer-related deaths worldwide, and non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer cases (Friedlaender et al., 2020; Hirsch et al., 2017). Even though great progresses have been achieved in the diagnosis and treatment of cancers, the 5-year survival rate of NSCLC patients is still low with <19% (Tsim et al., 2010). Therefore, the development of novel and efficient biomarkers for NSCLC is still necessary.
Long non coding RNAs (lncRNAs) are a class of long RNA transcript with more than 200 nucleotides in length (Jathar et al., 2017). It has been reported that lncRNAs can regulate the expression of downstream genes by acting as competing endogenous (ceRNAs) (Chan & Tay, 2018). To data, many lncRNAs have been identified to be closely associated with the development of NSCLC, such as pro-transition associated RNA (Yu et al., 2019), X-inactive specific transcript (Sun et al., 2017), Translation regulatory long non-coding RNA 1 (Luo et al., 2020). Forkhead box P4 antisense RNA 1 (FOXP4-AS1) is a recently identified lncRNA and has been studied in several human cancers including colorectal cancer (Li et al., 2017), gastric cancer (Chen et al., 2019), and cervical cancer (Zhao et al., 2020). However, the role of FOXP4-AS1 in NSCLC remains unclear.
Increasing evidence has indicated that lncRNAs can participate in the development of human cancers through interacting with microRNAs (miRNAs), another type of non-coding RNA with ∼22 nucleotides in length (Qi et al., 2015). One previous study reported that miR-3184-5p was a target of FOXP4-AS1, and FOXP4-AS1 promoted the progression of prostate cancer by inhibiting miR-3184-5p (Wu et al., 2019). This study aimed to investigate the function of FOXP4-AS1 and miR-3184-5p in NSCLC.
Eukaryotic translation initiation factor 5A (EIF5A), an 18-kDa protein, is essential for cell proliferation (Mathews & Hershey, 2015). EIF5A has been identified as a new oncogene in some types of human cancer including gastric cancer (Meng et al., 2015), glioblastoma (Preukschas et al., 2012), colorectal carcinoma (Zhu et al., 2012), and lung adenocarcinoma (Chen et al., 2002). Previous studies demonstrated that low expression levels of EIF5A was closely involved in the reductive progression including epithelial-mesenchymal transition, migration, invasion and proliferation of NSCLC cells (Xu et al., 2013), indicating that EIF5A was also an oncogene in NSCLC.
In this study, we demonstrated that FOXP4-AS1 was upregulated and miR-3184-5p was downregulated in NSCLC. Downregulation of FOXP4-AS1 obviously prevented the progression of NSCLC both in vitro and in vivo by regulating the miR-3184-5p/EIF5A axis. Our study provided a potential therapeutic target for NSCLC.
2 MATERIALS AND METHODS
2.1 Clinical samples
A total of 30 pairs of NSCLC tissue samples and matched adjacent normal tissue samples were collected from NSCLC patients by surgical resection at Kunming Yan'an Hospital between January 2017 to December 2019. The clinical samples were stored at −80°C for the subsequent analyses. This study was approved by the Human Ethics Review Committee of Kunming Yan'an Hospital. All participants provided their informed consent.
2.2 Cell culture
Human NSCLC cell lines (A549 [CCL-185], Calu-1 [HTB-54] and H1299 [CRL-5803]) and normal bronchial epithelial cell line human bronchial epithelial cells (CRL-2078) were obtained from american type culture collection. Cells were cultured in RPMI1640 medium (Gibco) supplemented with 10% fetal bovine serum in a 37°C incubator with 5% CO2.
2.3 Cell transfection
Two small interfering RNAs targeting FOXP4-AS1 (si-FOXP4-AS1 #1 and si-FOXP4-AS1 #2), miR-3184-5p mimics/inhibitor and the corresponding negative controls (miR-NC and inhibitor NC) were purchased from Genepharma. To overexpress EIF5A, the full length of EIF5A was amplified and cloned into the expression vector pcDNA3.1 and named pc-EIF5A. The empty vector was used as the negative control (vector). Next, 50 nM siRNAs/mimics/inhibitor or recombinant expression vector were transfected into A549 and Calu-1 cells using Lipofectamine 3000 (Invitrogen) as previously described (Lu & Zhang, 2019).
2.4 Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNAs were extracted from tissues and cultured cells using the TRIzol method. The cDNA was synthesized using a reverse transcription kit (Invitrogen), and PCR reactions were performed on PCR iCycler device (Bio-Rad) using Synergy Brands Green PCR kit (Toyobo, Japan). The relative expression of targets was calculated using the 2−ΔΔCt method (Arocho et al., 2006) with U6 and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal references. The primers used were listed in Table 1.
| Gene name | RefSeq and product length | Primer sequence (5′ to3′) |
|---|---|---|
| lncFOXP4-AS1 | NC_000006.12 | Forward: 5′-ATGGAAAAGGGGCTGACTGG-3′ |
| 195 bp | Reverse: 5′-CTCTGGGAATGGGGTGTACG-3′ | |
| miR-3184-5p | NC_000017.11 | Forward: 5′-TGAGAAACCTCAGATTGAGCTTTT-3′ |
| 75 bp | Reverse: 5′-CTCTACAGCTATATTGCCAGCCA-3′ | |
| EIF5A | NC_000017.11 | Forward: 5′-GAGCAGAAGTACGACTGTGGAG-3′ |
| 142 bp | Reverse: 5′-CAGGTTCAGAGGATCACTGCTG-3′ | |
| GAPDH | NC_000012.12 | Forward: 5′-ATCCACGGGAGAGCGACAT-3′ |
| 101 bp | Reverse: 5′-CAGCTGCTTGTAAAGTGGAC-3′ | |
| U6 | NC_000006.12 | Forward: 5′-ACAGATCTGTCGGTGTGGCAC-3′ |
| 182 bp | Reverse: 5′-GGCCCCGGATTATCCGACATTC-3′ |
- Abbreviation: GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
2.5 Western blot
Total proteins were isolated from tissues and cultured cells using radioimmunoprecipitation buffer. After separating by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferring into polyvinylidene fluoride membranes, the membranes were incubated with rabbit monoclonal antibodies anti-EIF5A (1:1000, Thermo Fisher Scientific) and anti-GAPDH (1:1000, Invitrogen) at 4°C overnight. The membranes were then subjected to the secondary HRP-conjugated antibody (1:1000, Invitrogen). The protein bands were observed using enhanced chemiluminescence substrates.
2.6 Colony formation assay
Colony formation assay was conducted as previously described (Xu et al., 2021). Briefly, approximately 1000 A549 and Calu-1 cells were seeded into 6-well plates and cultured for 2 weeks. Cells were then stained with 0.1% crystal violet solution. The numbers of colonies were counted and photographed under a light microscope.
2.7 Flow cytometry
Cell apoptosis was evaluated using the Annexin V-fluorescein isothiocyanate (fluorescein isothiocyanate (FITC)) apoptosis detection kit (Beyotime) as previously described (Chai et al., 2021). Briefly, 5 × 105 A549 and Calu-1 cells were incubated with 10 μl FITC and 5 μl propidium iodide for 15 min. Finally, the percentage of apoptotic cells was calculated using FACSCalibur (BD Biosciences).
2.8 Luciferase reporter assay
The target prediction was performed using the online database (https://starbase.sysu.edu.cn/, http://bioinformatics.mdanderson.org/main/TANRIC:Overview). The pmirG1O dual-luciferase reporter vectors containing the wild type (WT) or mutant (MUT) of FOXP4-AS1 and 3′-UTR of EIF5A against the miR-3184-5p binding site were constructed by Genepharma. MiR-3184-5p mimics or miR-NC was transfected into 293T cells using Lipofectamine 3000. After transfection for 48 h, the relative luciferase activity was detected.
2.9 In vivo animal model
The in vivo animal model was constructed using BALB/C (nu-nu) nude mice (approximately 5 weeks old and weighted 18 g) as previously described (Yang et al., 2019). Firstly, knockdown of FOXP4-AS1 was achieved by transfecting A549 cells with short hairpin RNA targeting FOXP4-AS1 (sh-FOXP4-AS1), and puromycin was used to screen stably transfected cell line as previously described (Guo & Li, 2019). To construct the in vivo animal model, approximately 3 × 106 stably transfected A549 cells were subcutaneously injected into the nude mice. Tumor volume was evaluated every 5 days for a total of 20 days according to the formula: volume = 1/2 × length × width2. On the 20th day, the tumors were removed, imaged and weighted. Subsequently, the tumor tissues were cut into 4-μm sections, and incubated with anti-Ki67 antibody (1:200, Abcam) and anti-EIF5A antibody (1:200, Abcam) at 4°C overnight. Then the sections were subjected to biotinylated secondary antibodies to incubate for 1 h. Immunohistochemistry images were captured by an Olympus microscope. This study was approved by the Animal Ethics Committee of Kunming Yan'an Hospital.
2.10 Statistical analysis
Data were presented as mean ± standard deviation (SD) and analyzed using GraphPad Prism 7. The difference between two groups was tested by Student's t-test, and differences among multiple groups were determined by one-way ANOVA. p < 0.05 was considered as statistically significant.
3 RESULTS
3.1 FOXP4-AS1 was significantly upregulated and miR-3184-5p was downregulated in NSCLC
We firstly detected the expression of FOXP4-AS1 and miR-3184-5p in NSCLC tissues and cell lines. The results showed that FOXP4-AS1 was significantly upregulated and miR-3184-5p was downregulated in NSCLC tissues compared with that in matched adjacent normal tissues (Figure 1a,b). Meanwhile, FOXP4-AS1 was also significantly upregulated and miR-3184-5p was downregulated in three NSCLC cell lines (Figure 1c,d). These results suggested that FOXP4-AS1 and miR-3184-5p might play important roles in NSCLC progression.
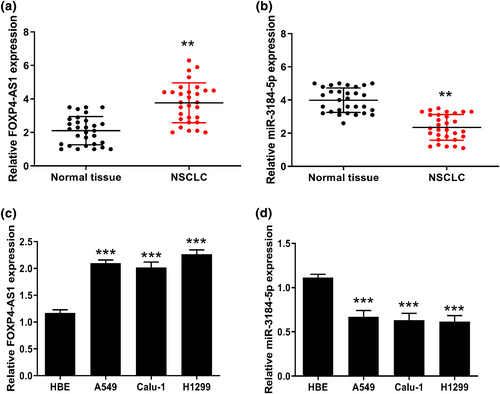
The expression of FOXP4-AS1 and miR-3184-5p in non-small cell lung cancer (NSCLC). (a and b) The expression of FOXP4-AS1 (a) and miR-3184-5p (b) in NSCLC tissues and matched adjacent normal tissues was evaluated by qRT-PCR (n = 30). (c and d) The expression of FOXP4-AS1 (c) and miR-3184-5p (d) in NSCLC cell lines was evaluated by qRT-PCR. **p < 0.01, and ***p < 0.001
3.2 Downregulation of FOXP4-AS1 reduced the proliferation and induced apoptosis of NSCLC cells
Then, two siRNAs targeting FOXP4-AS1 were transfected into A549 and Calu-1 cells, and the transfection efficiency was confirmed by qRT-PCR (Figure 2a). Cell proliferation was evaluated and the results showed that downregulation of FOXP4-AS1 significantly reduced the viability and colony formation of two cell lines compared with si-NC (Figure 2b,c). In addition, flow cytometry results showed that downregulation of FOXP4-AS1 significantly promoted cell apoptosis of two cell lines compared with si-NC (Figure 2d). These results indicated that downregulation of FOXP4-AS1 reduced the proliferation and induced apoptosis of NSCLC cells.
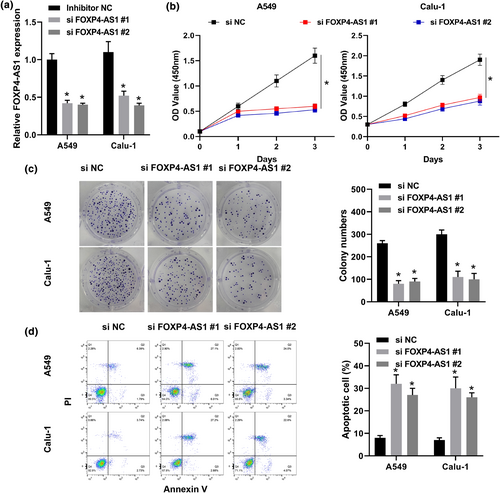
The effect of FOXP4-AS1 on the proliferation and apoptosis of non-small cell lung cancer cells. A549 and Calu-1 cells were transfected with si-FOXP4-AS1 #1 and si-FOXP4-AS1 #2. (a) The expression of FOXP4-AS1 was evaluated by qRT-PCR. (b) CCK-8 assay. (c) Colony formation assay. (d) Flow cytometry assay. *p < 0.05
3.3 FOXP4-AS1 regulated EIF5A by binding to miR-3184-5p
To confirm the binding relationship between FOXP4-AS1 and miR-3184-5p, Starbase was used to predict the binding site (Figure 3a). Luciferase reporter assay was conducted in 293T cells and the results showed that miR-3184-5p mimics significantly reduced the relative luciferase activity of FOXP4-AS1 WT compared with miR-NC, but exhibited no effect on FOXP4-AS1 MUT (Figure 3b). Meanwhile, downregulation of FOXP4-AS1 significantly increased the expression levels of miR-3184-5p in A549 and Calu-1 cells compared with si-NC (Figure 3c). Furthermore, the potential targets of miR-3184-5p were predicted by Starbese and it showed that miR-3184-5p might bind to the 3′-UTR of EIF5A (Figure 3d), and luciferase reporter assay revealed that miR-3184-5p mimics significantly reduced the relative luciferase activity of WT 3′-UTR EIF5A compared with miR-NC, but had no effect on MUT 3′-UTR EIF5A (Figure 3e). In addition, we evaluated the effect of FOXP4-AS1 on the expression of EIF5A, and the results showed that downregulation of FOXP4-AS1 significantly reduced the expression levels of EIF5A at both mRNA and protein levels in A549 and Calu-1 cells compared with si-NC (Figure 3f,g). These results suggested that FOXP4-AS1 regulated EIF5A possibly by binding to miR-3184-5p in NSCLC.
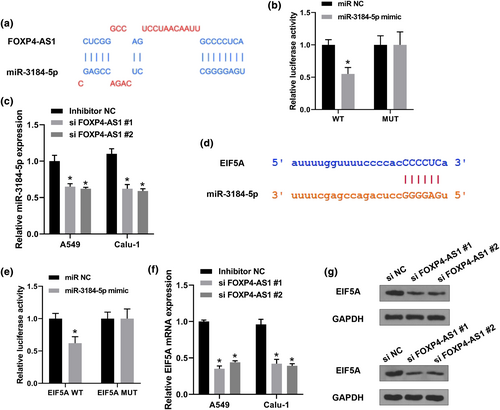
FOXP4-AS1 regulated EIF5A though binding to miR-3184-5p in non-small cell lung cancer (NSCLC). (a) The binding site between FOXP4-AS1 and miR-3184-5p was predicted by Starbase. (b) Luciferase reporter assay. (c) A549 and Calu-1 cells were transfected with si-FOXP4-AS1 #1 and si-FOXP4-AS1 #2. The expression of miR-3184-5p was detected by qRT-PCR. (d) The putative binding site between miR-3184-5p and EIF5A was predicted by Starbase. (e) Luciferase reporter assay. (f and g) A549 and Calu-1 cells were transfected with si-FOXP4-AS1 #1 and si-FOXP4-AS1 #2. The expression of EIF5A was evaluated by qRT-PCR (f) and western blot (g). *p < 0.01
3.4 Downregulation of miR-3184-5p reversed the inhibitory effects of knockdown of FOXP4-AS1 on NSCLC cell proliferation and apoptosis
To further determine the functions of FOXP4-AS1 and miR-3184-5p in NSCLC, A549 and Calu-1 cells were co-transfected with si-FOXP4-AS1 and miR-3184-5p inhibitor. It showed that miR-3184-5p inhibitor + si-NC significantly increased cell viability and the number of colonies compared with that in the si-NC + miR-NC group; si-FOXP4-AS1 + miR-NC significantly reduced cell viability and the number of colonies compared with that in the si-NC + miR-NC group; while si-FOXP4-AS1 + miR-3184-5p inhibitor obviously attenuated the inhibitory effects of si-FOXP4-AS1 + miR-NC on cell viability and colony formation (Figure 4a,b). In addition, si-FOXP4-AS1 + miR-3184-5p inhibitor significantly reduced apoptotic rate of A549 and Calu-1 cells transfected with si-FOXP4-AS1 + miR-NC (Figure 4c). These results indicated that downregulation of miR-3184-5p reversed the inhibitory effects of si-FOXP4-AS1 on proliferation and apoptosis of NSCLC cells.
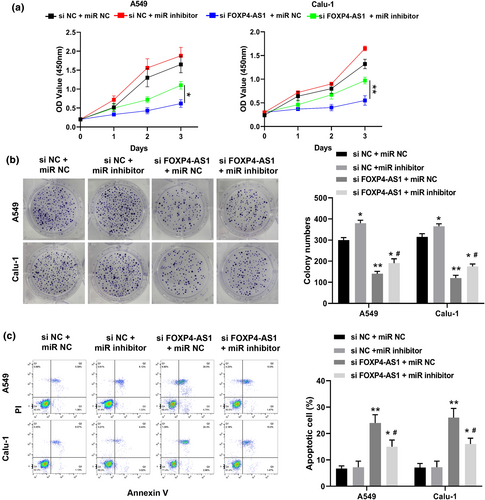
Downregulation of miR-3184-5p reversed the inhibitory effects of si-FOXP4-AS1 on proliferation and apoptosis of non-small cell lung cancer cells. A549 and Calu-1 cells were co-transfected with si-NC and miR-NC, si-NC + miR-3184-5p inhibitor, si-FOXP4-AS1 + miR-NC, or si-FOXP4-AS1 and miR-3184-5p inhibitor. (a) CCK-8 assay. (b) Colony formation assay. (c) Flow cytometry assay. *p < 0.05 and **p < 0.01 versus si-NC + miR-NC group or si-FOXP4-AS1 + miR-NC group; #p < 0.05 versus si-FOXP4-AS1 + miR-NC group
3.5 Overexpression of EIF5A reversed the inhibitory effects of si-FOXP4-AS1 on the proliferation and apoptosis of NSCLC cells
To confirm that EIF5A mediated the role of FOXP4-AS1, A549 and Calu-1 cells were co-transfected with si-FOXP4-AS1 and pc-EIF5A. We found that pc-EIF5A + si-NC significantly increased the viability and colonies of these two cell lines compared with that in the vector + si-NC group; co-transfection of pc-EIF5A and si-FOXP4-AS1 obviously attenuated the inhibitory effects of si-FOXP4-AS1 + vector on cell viability and colony formation (Figure 5a,b). Meanwhile, apoptosis assay showed that co-transfection of pc-EIF5A and si-FOXP4-AS1 also significantly reduced the apoptotic rate of A549 and Calu-1 cells transfected with si-FOXP4-AS1 + vector (Figure 5c). These results indicated that overexpression of EIF5A reversed the inhibitory effects of si-FOXP4-AS1 on proliferation and apoptosis of NSCLC cells.
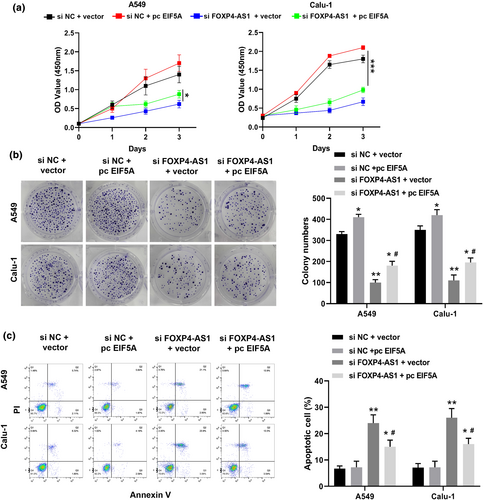
Overexpression of EIF5A reversed the inhibitory effects of si-FOXP4-AS1 on proliferation and apoptosis of non-small cell lung cancer cells. A549 and Calu-1 cells were co-transfected with si-NC and vector, si-NC + pc-EIF5A, si-FOXP4-AS1 + vector, or si-FOXP4-AS1 + pc-EIF5A. (a) CCK-8 assay. (b) Colony formation assay. (c) Flow cytometry assay. *p < 0.05 and **p < 0.01 versus si-NC + vector group or si-FOXP4-AS1 + vector group; #p < 0.05 versus si-FOXP4-AS1 + vector group
3.6 Knockdown of FOXP4-AS1 inhibited tumor growth in vivo
Finally, we evaluated the role of FOXP4-AS1 in NSCLC progression in vivo. The results showed that knockdown of FOXP4-AS1 significantly inhibited tumor volume and tumor weight compared with that in the negative control sh-NC (Figure 6a–c). Meanwhile, immunohistochemistry assay showed that knockdown of FOXP4-AS1 obviously reduced the expression levels of EIF5A and Ki67 in tumor tissues compared with sh-NC (Figure 6d). These results suggested that knockdown of FOXP4-AS1 inhibited tumor growth in vivo. In addition, the related inflammatory indicators (IL-1β, IL-6, and TNF-a) were detected in sh-NC or sh-FOXP4-AS1 transfected cells. The results showed that the effect of knockdown of FOXP4-AS1 on the inflammatory markers was not statistically significant (Supplementary File).
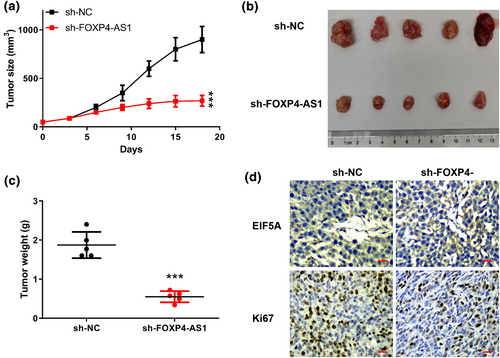
Knockdown of FOXP4-AS1 inhibited tumor growth in vivo. (a) Tumor volume. (b) The representative images of tumors from two groups. (c) Tumor weight. (d) The expression of EIF5A and Ki67 in tumor tissues was evaluated by immunohistochemistry assay. Scale bar = 100 μm, magnification ×200. ***p < 0.001
4 DISCUSSION
In the present study, we found for the first time that FOXP4-AS1 was highly expressed in NSCLC tissues and cell lines. Furthermore, downregulation of FOXP4-AS1 significantly inhibited cell proliferation and promoted apoptosis of the two NSCLC cell lines, confirming the oncogenic role of FOXP4-AS1. Interestingly, miR-3184-5p was reported to be a direct target of FOXP4-AS1 (Wu et al., 2019), and our results revealed that miR-3184-5p was downregulated in NSCLC tissues and cancer cell lines. Meanwhile, luciferase reporter assay revealed the binding relationship between FOXP4-AS1 and miR-3184-5p. Moreover, downregulation of miR-3184-5p significantly reversed the inhibitory effects of si-FOXP4-AS1 on cell proliferation and apoptosis. These data further determined the regulatory role of FOXP4-AS1 and miR-3184-5p in NSCLC progression. Significantly, increasing evidence has suggested that lncRNAs have different roles in cancer progression and tumorigenesis and are widely expressed in lung carcinoma (Lu et al., 2018). LncRNAs can serve as new therapeutic targets for NSCLC by restoring the specificity and sensitivity of cancer cells to chemotherapeutic drugs and they may even cure the condition due to their effects on the development and regulation of diverse molecular pathways correlated with gene expression (Ricciuti et al., 2016). This study extended our understanding of the FOXP4-AS1/miR-3184-5p axis in the development of NSCLC, and provided that FOXP4-AS1 might be a potential therapeutic target.
This study aimed to investigate the functions of FOXP4-AS1/miR-3184-5p in NSCLC. Besides miR-3184-5p, some other miRNA targets of FOXP4-AS1 have been identified in human diseases. For example, high expression levels of FOXP4-AS1 predicts a poor prognosis and attenuates the development of mantle cell lymphoma through regulating the miR-423-5p/NACC1 pathway (Tao et al., 2021). FOXP4-AS1 is associated with cervical cancer progression by targeting the miR-136-5p/CBX4 axis (Zhao et al., 2020). Whether these known miRNA targets or other un-identified miRNA targets mediated the function of FOXP4-AS1 in NSCLC progression need to be further explored. These findings will increase our knowledge involved in the pathogenesis of NSCLC.
MiRNAs have been reported to regulate the expression of downstream genes by binding to the 3′-UTR of their targets (Mohr & Mott, 2015). We found that EIF5A was a potential target of miR-3184-5p. Luciferase reporter assay also confirmed this prediction. Moreover, rescue experiments were performed and indicated that overexpression of EIF5A obviously reversed the effects of si-FOXP4-AS1 on NSCLC progressions including cell proliferation and apoptosis. In addition, in previous studies, there were also several target genes that were shown to mediate the function of miR-3184-5p in human diseases. For example, miR-3184-5p attenuates neuropathic pain through targeting SCN3A in chronic constriction injury (Ye et al., 2020). MiR-3184-5p is closely associated with the progression of cervical cancer by negatively regulating Erb-B2 Receptor Tyrosine Kinase 2 (Liu et al., 2021). MiR-3184-5p also participates in proliferation of esophageal squamous cell carcinoma cells by targeting FOXP4 (Li et al., 2020). MiRNAs have been shown to target multiple downstream genes simultaneously (Lee & Dutta, 2009). Whether one or more target genes participated in the function of miR-3184-5p in NSCLC should be determined in our subsequent experiments in the future.
However, limitations exist in the present study: if we manipulated the expression levels of miR-3184-5p or EIF5A in animal model in vivo, the tumor growth of NSCLC affected by FOXP4-AS1 might be changed. We will address these limitations, which contribute to confirm the role of FOXP4-AS1/miR-3184-5p/EIF5A axis in NSCLC progression.
5 CONCLUSION
In summary, we demonstrated that FOXP4-AS1 promoted the progression of NSCLC by targeting the miR-3184-5p/EIF5A axis, suggesting that FOXP4-AS1 might be a potential therapeutic target.
ACKNOWLEDGMENT
Not Applicable.
CONFLICT OF INTEREST
All other authors have no conflicts of interest. We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
AUTHOR CONTRIBUTIONS
Dingbiao Li, Zhenhua Li: study concepts, literature research, clinical studies, data analysis, experimental studies, manuscript writing and review; Wang YanFei, Ying Wang: study design, literature research, experimental studies and manuscript editing; Jianlin Shi, Chang Liu: definition of intellectual content, clinical studies, data acquisition and statistical analysis; Laihao Qu, Shoujun Deng: data acquisition, manuscript preparation and data analysis; Dalin Xiong: data acquisition and statistical analysis. All authors have read and approve the submission of the manuscript.
CONSENT TO PUBLISH
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to their containing information that could compromise the privacy of research participants.




