Ecological and evolutionary inferences from aphid microbiome analyses depend on methods and experimental design
Abstract
Introduction
Aphids play an important role in agroecological contexts as pests and vectors of plant diseases. Aphid performance is closely connected to microbial endosymbionts that provide different benefits or costs to both the aphids and their hosts plants. Furthermore, the microbiome of aphids is connected to soil microbiomes via the plant. Aphid microbiome experiments usually include a pooling step, where several individuals are sequenced together to obtain sufficient DNA concentrations but pooling may blur intraspecific variations.
Materials and Methods
To investigate the effects of sequencing single versus pooled aphids on the results of microbiome analyses, we compared 16S rRNA/ITS amplicon libraries from pooled and single oak aphids (Tuberculatus annulatus HARTIG) under three different soil treatments. We tested whether results quantitatively or qualitatively depend on pooling aphids, prevalence-based in silico filtering or removal of the primary endosymbiont (Buchnera aphidicola). Buchnera phylogeny, prevalence and abundance of secondary endosymbionts and effects of soil microbiota were investigated.
Results
Pooling leads to quantitative differences in bacteria and qualitative differences in fungal species richness, bacterial community composition and partially fungal community composition. Filtering-dependent results were obtained for bacterial evenness. Buchnera phylogeny supports the hypothesis of cospeciation of primary endosymbionts in oak aphids. We detected Arsenophonus, Hamiltonella, Rickettsia, Rickettsiella, Serratia and Sphingopyxis in oak aphids, with their prevalence and abundance partially affected by pooling. Pooling leads to overestimating the frequency of multispecies endosymbiont infections, while underestimating their relative abundance.
Conclusion
We hereby extend our view on non-model aphid microbiomes and identify pitfalls in experimental design in aphid microbiome research.
1 INTRODUCTION
Aphids are of major concern for current agriculture. They damage crops and reduce yield by ingesting plant sap, injecting potentially phytotoxic saliva, vectoring plant diseases and indirectly decreasing photosynthetic rate via the exudation of honeydew that is subsequently colonized by sooty moulds (Dedryver et al., 2010; Simon & Peccoud, 2018). Aphid species found in agricultural settings are favoured by the combination of temporal habitat instability combined with the spatial habitat uniformity realized in agroecosystems (Frantz et al., 2006; Gilabert et al., 2009; Yan et al., 2020). Apart from cyclical parthenogenesis (alternating sexual and asexual life stages) and phenotypic plasticity (ability to express different phenotypes under environmental changes), the metabolic relationship of aphids with microbial symbionts is important for their adaptive potential (Simon & Peccoud, 2018) and crucial for their survival. Plant sap constitutes a one-sided diet, deficient in essential nutrients that the aphid requires for survival and development, for example, essential amino acids or vitamins (Baumann, 2005; Moran & Baumann, 2000). Bacterial endosymbionts in aphids are categorized into primary, obligate endosymbionts and secondary, facultative endosymbionts. Research on aphid endosymbionts usually focuses on aphid species that are polyphagous and/or feed on important crops, for example, Myzus persicae SULZER or Acyrtosiphon pisum HARRIS. Only a few studies focus on oligophagous aphid species and the response of their associated microbial symbionts but these could be affected by geographical distribution and climatic conditions (Xu et al., 2021). Insights in oligophagous aphid symbiont communities could both challenge or support our current view on aphid microbiomes.
Aphid microbiome sampling is usually performed by pooling several individuals to obtain sufficient amounts of microbial DNA. Depending on the research question, pooling may not be appropriate for investigating aphid-associated microbes, considering that intraspecific variations may not be properly represented in the data set. Using suitable laboratory protocols, DNA extraction of both bacterial (Jousselin et al., 2016; Xu et al., 2021) and fungal DNA (data set of 10) from a single individual is possible. On the other hand, samples with low microbial biomass like in single aphids are sensitive to contaminants during processing (Caruso et al., 2019). Contaminant DNA arising from DNA extraction, sequencing and inconsistent or slightly differently performed handling can have strong effects on finally obtained data, where the contaminants can even outcompete the biological signal within samples (Caruso et al., 2019; Eisenhofer et al., 2019; Jousselin et al., 2016). The subtractive removal of contaminant reads in silico using negative controls (Davis et al., 2018), or exclusion of ASVs with low abundance or frequency (Cao et al., 2021) can be done to account for sequence contamination. However, the exclusion of rare or low-abundance taxa results in an incomplete taxonomic and functional profile of the microbiome (Banerjee et al., 2018). Therefore, excessive data cleaning may change ecological inferences regarding the prevalence of rare taxa despite their potentially crucial ecosystem functions (Banerjee et al., 2018; Jousset et al., 2017; Reid et al., 2011; Schloss, 2020) and will also quantitatively affect diversity indices (Cao et al., 2021; Reitmeier et al., 2021; Schloss, 2020).
Plant sap-sucking insects like aphids heavily depend on microbial endosymbionts to supplement their one-sided diet with vital nutrients. The interdependence of aphids and their primary endosymbiont (usually the bacterium Buchnera aphidicola, ad Enterobacteriaceae) resulted in congruent phylogenies of host and symbiont (Baumann, 2005; Martinez-Torres et al., 2001). Thus, the genome of the bacterial endosymbionts can be used to resolve the phylogenetic relationship of their host in aphids (Martinez-Torres et al., 2001; Nováková et al., 2013), a phenomenon allegeable by symbiotic cospeciation. For cospeciation analyses however, the endosymbiont sequence should be as reliable and representative for a given aphid species as possible because these phylogenetic analyses will be easily influenced by single nucleotide polymorphisms. In amplicon analyses, we often obtain several ASVs assigned to the same microbial species. These ASVs can be interpreted as representing the intraspecific genetic diversity of this microbial species, but they could also simply derive from sequencing or read-joining errors. Random sequencing errors can be accounted for by using operational taxonomic units (OTUs) with a 97% similarity cut-off for statistical analyses, but this method holds the disadvantage of potentially underestimating the genetic diversity of the sample compared to the usage of ASVs. Other statistical tools often used in metagenomic analyses, for example, DADA2 (Callahan et al., 2016), perform by default sequencing error filtering based on prevalence, read length, base pair quality scores and/or other methods. For amplicon libraries, the questions are if a 300 bp amplicon provides sufficient information to construct phylogenetic trees for the aphid host based on Buchnera-assigned ASVs and if all such ASVs in a given aphid microbiome data set would support the current concept of cospeciation in aphids.
Secondary endosymbionts (secES) in aphids are facultative microbial endosymbionts that are usually horizontally transmitted but can provide several important ecological functions to their host. Known secES in aphids belong to Alphaproteobacteria and Gammaproteobacteria. They act partially as a backup or complement for the metabolic pathways provided by the genetically highly genome-degraded primary endosymbiont (Manzano-Marı́n et al., 2020, 2023) or mediate protection to the aphid host against biological threats like pathogens or parasitoids (Guo et al., 2017; Scarborough et al., 2005; Zytynska & Weisser, 2016). Despite research in aphid secES often focuses on polyphagous aphid species that occur as agricultural pests, protection against abiotic and biotic stressors is also important for survival in oligophagous aphid species. Since secES can negatively affect aphid fecundity, the net fitness benefits of housing an additional endosymbiont depend on the ecological context of the aphid host (Guo et al., 2017; Zytynska et al., 2021), for instance, parasitoid abundance or climatic conditions. Therefore, secES abundance and prevalence in an aphid colony may be low under non-stress conditions, only increasing if the respective environmental stressor appears. As a facultative part of the aphid microbiome, secES abundance and prevalence could be underestimated if aphids are pooled in the course of an experimental setup, as well as when using in silico filtering, on both individual aphid and population level.
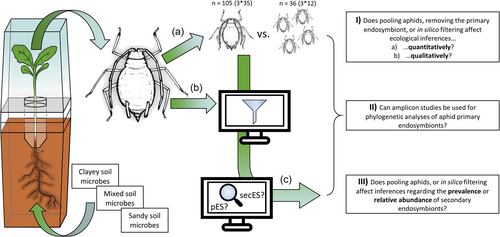
This article aims to identify potential methodological pitfalls in designing and performing amplicon analyses of aphids. Therefore, it is intended to cover several different aspects of aphid microbiome analyses that need to be considered for future research questions and experimental designs. Further, the analysis deepens our understanding of the microbiome associated with the common oak aphid (Tuberculatus annulatus HARTIG), an oligophagous aphid species specialized in oak and partially found on chestnuts (Castanea spp.) (Dransfield & Brightwell, 1999). We reanalysed a recently published amplicon data set (Wolfgang et al., 2023), containing single and pooled oak aphids infesting oak seedlings grown in different soil microbiomes (Figure 1). The data set was used to investigate the effects of soil microbiota on aphid microbiomes in a plant-mediated way before (Wolfgang et al., 2023). We here evaluate whether we can obtain the same inferences when using single or pooled aphid samples, and/or using different in silico filtering approaches. The data set includes bacterial and fungal data and was used to answer the following questions: (I) Does pooling, prevalence-based filtering or primary endosymbiont removal quantitatively or qualitatively affect ecological inferences on aphid microbiomes? (II) Can ASVs derived from amplicon studies be used for phylogenetic analyses of aphid primary endosymbionts? (III) Does pooling aphids, or in silico filtering affect inferences regarding the prevalence or relative abundance of secES?
2 MATERIALS AND METHODS
2.1 Experimental design
We reinvestigated the full data set of Wolfgang et al. (2023), including samples of single and pooled (four specimens) aphids. Data is publicly available at the European Nucleotide Archive repository under accession number PRJEB50358. Shortly, this experiment included pedunculate oak (Quercus robur L.) seedlings in microcosms that separate above- and below-ground seedling compartments to prevent cross-contamination between soil and phyllosphere (Abdelfattah, 2021; Abdelfattah et al., 2021). Acorns were planted in three soils that were physicochemically standardized but included different natural soil inocula (microbiota originating from either clayey, mixed or sandy soil type). Seven-day-old oak seedlings were infested with 20 individuals of common oak aphids (T. annulatus HARTIG), and after another 7 days, soil, phyllosphere and aphids were sampled, and metagenomic DNA was extracted. A total of 97 pooled aphid samples were obtained for clayey, mixed and sandy soil treatment, respectively (3 × 35 replicates, minus eight samples removed because of visible disease symptoms of the plants), all originating from different plants. In addition, 36 single aphids (12 specimens per soil treatment, respectively) were processed. The data set further includes real-time quantitative PCR results for pooled (nbac = 11, nfun = 12) and single (nbac = 12, nfun = 11) aphids. The amplicon samples were sequenced using 16SrRNA and ITS amplicon sequencing on a MiSeq V3 (600-cycle) platform for 300 bp paired-end sequencing. For bacteria, a PCR product could be amplified in all samples, while for fungi, a PCR product could be obtained in 92% (89 of 97 samples) and 47% (17 of 36 samples) of the samples for pooled and single aphid samples, respectively. Demultiplexing was performed in QIIME v. 2019.10 (Bolyen et al., 2019) using cutadapt (Martin, 2011), truncated at 150 bp (bacteria) and 170 bp (fungi) and denoised using DADA2 (Callahan et al., 2016). The phylogenetic tree was generated using mafft (Katoh, 2002) and fasttree2 (Price et al., 2010). Subsequent statistical analyses were performed in R version 4.1.1 (R Core Team, 2018) using phyloseq v. 1.38.0 (McMurdie & Holmes, 2013) and microViz v. 0.10.6 (Barnett et al., 2021). Contaminant ASVs were identified and removed using a prevalence-based method implemented in the ‘decontam’ package v. 1.14.0 (Davis et al., 2018). Sequencing depth was compared using raw single and pooled read counts using t-test, rarefaction curves were generated using ‘ranacapa’ v. 0.1.0 (Kandlikar et al., 2018) (Supporting Information S1: Figure 1). All statistical analyses were performed using (a) an unfiltered data set, (b) a Buchnera-filtered data set, (c) a data set only containing core taxa using a 16% prevalence cut-off and (d) a Buchnera-filtered data set only containing core taxa using a 16% prevalence cut-off. The 16% prevalence refers to a minimum prevalence of 50% of an ASV that would be unique to one of the three soil treatments. For alpha diversity indices (species richness, evenness, Shannon diversity) estimation, data sets were rarefied to 1500 (bacteria unfiltered), 1000 (bacteria unfiltered without Buchnera, bacteria filtered, fungi unfiltered and fungi filtered) and 500 sequences (bacteria filtered without Buchnera) based on retaining a maximum number of samples (Supporting Information S1: Table 1). For abundance, the raw read counts were adjusted according to the percentage of reads removed by Buchnera- or prevalence-based filtering in amplicon samples. Beta diversity analyses (Bray−Curtis, weighted UniFrac) were performed on CSS-transformed data sets.
2.2 Quantitative effects of pooling and filtering on diversity indices (Ia)
Microbial alpha diversity indices between all pooled and all single aphid samples were compared, irrespective of soil treatment. Sequencing depth significantly higher in pooled aphids samples (t-test bacteria: p = 0.038; fungi: p = 0.025). We assessed the quantitative effects of pooling on aphid microbiome species richness, Pielou's evenness, Shannon diversity, abundance based on qPCR results [16SrRNA and ITS for bacteria and fungi, respectively (Wolfgang et al., 2023)] and community structure based on Bray−Curtis dissimilarities and weighted UniFrac distances. Data for species richness, Pielou's evenness, Shannon diversity, abundance and community composition were modelled as a function of ‘pooling’, using linear models for alpha diversity indices and abundance, and PERMANOVA for community composition.
2.3 Qualitative effects of pooling and filtering on ecological inferences (Ib)
Microbial diversity indices in aphids were modelled as a function of soil treatment separately for pooled and single aphids. Differences in species richness, Pielou's evenness, Shannon diversity and abundance were modelled using ANOVA with soil treatment as the independent variable. Differences in microbial community composition based on Bray−Curtis dissimilarities were modelled as a function of soil treatment using PERMANOVA, and compared pairwise using the 'pairwiseAdonis' package (Martinez Arbizu, 2017).
2.4 Buchnera phylogeny in T. annulatus (II)
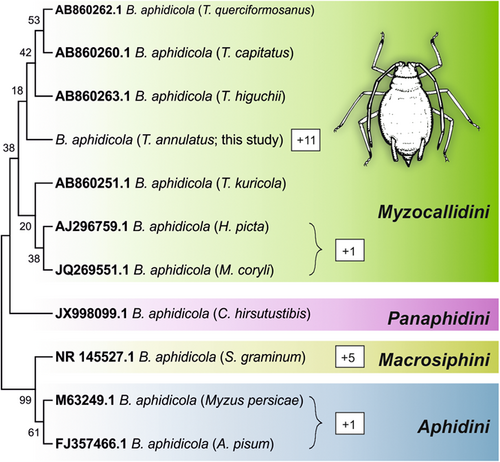
ASVs assigned to Buchnera were extracted from the data set and aligned using MUSCLE (Edgar, 2004) implemented in MEGA 11 (Tamura et al., 2021). Aligned sequences were trimmed to the same length. The most abundant and prevalent ASV assigned to Buchnera was identified as the representative haplotype and supplemented with sequences of B. aphidicola originating from other aphid host species available at NCBI (https://www.ncbi.nlm.nih.gov) (Figure 1). A neighbour-joining tree was constructed in MEGA 11 using the standard settings including the bootstrap method (n = 1000) to assess statistical confidence. The phylogenetic tree construction steps were repeated including the nonrepresentative Buchnera ASVs to identify any divergences in positioning of these ASVs in the phylogenetic tree.
2.5 Secondary endosymbiont abundance and prevalence in T. annulatus (III)
The Buchnera-filtered data set was checked on genus level for 10 taxa previously described as potential secES (Guo et al., 2017; McLean et al., 2019; Zytynska & Weisser, 2016), of which six genera were detected, namely Arsenophonus, Hamiltonella, Rickettsia, Rickettsiella, Serratia and Sphingopyxis. The data set was manually checked for the number of different secES found within one aphid sample. The prevalence of the six potential secES was assessed for pooled aphids, single aphids and the combined data set. To test whether the relative abundance in secES-positive aphid samples differs between pooled and single aphid samples, a Wilcoxon rank-sum test was performed for each secES separately.
3 RESULTS
3.1 Pooling and filtering can quantitively shift microbial diversity metrics (Ia)
Significant differences were observed in bacterial evenness and bacterial Shannon diversity, which was not dependent on filtering nor on Buchnera removal. For fungi, differences between pooled and single aphid microbial alpha diversity were not significant (Table 1). Significantly higher abundance based on qPCR reads in pooled aphids was only observed in two out of six data sets (Table 1). Prevalence-based ASV filtering increased the R2 value for pooled-single comparison in both Bray−Curtis dissimilarity and weighted UniFrac distances using PERMANOVA (0.11−0.2 vs. 0.29−0.37) (Supporting Information S1: Table 2). Both bacterial and fungal community composition significantly differed between single and pooled aphids, irrespective of data set filtering.
| Data set | Richness | Evenness | Shannon | Abundance | BC | wUF | |
|---|---|---|---|---|---|---|---|
| Bacteria | With Buchnera, raw | Single* | Single* | Single* | Pooled* | R2 = 0.013* | R2 = 0.012* |
| With Buchnera, filtered | Single* | Single* | Single* | Pooled* | R2 = 0.037* | R2 = 0.031* | |
| No Buchnera, raw | Single* | Single* | Single* | n.s. | R2 = 0.012* | R2 = 0.011* | |
| No Buchnera, filtered | Single* | Single* | Single* | n.s. | R2 = 0.035* | R2 = 0.029* | |
| Fungi | Raw | n.s. | n.s. | n.s. | n.s. | R2 = 0.020* | R2 = 0.018* |
| Filtered | n.s. | n.s. | n.s. | n.s. | R2 = 0.037* | R2 = 0.034* |
- Note: All analyses were performed with an unfiltered data set (‘raw’) and a filtered data set using a prevalence-based 16% threshold (‘filtered’); for bacteria, analyses were repeated using data sets with and without primary endosymbiont Buchnera aphidicola. Only the group with significantly (t-test, p < 0.05) higher diversity value is displayed. Abundance based on log10-transformed 16SrRNA (bacteria) and ITS (fungi) reads derived from qPCR. Abundance read filtering (Buchnera removal and prevalence-based filtering) was performed by subtracting the ratio that was removed by filtering in the respective amplicon sample from raw reads. R2 values for Bray−Curtis dissimilarity (BC) and weighted UniFrac (wUF) distance results based on PERMANOVA, asterisk indicate p-value below 0.05 (for detailed results, see Supporting Information S1: Table 2).
3.2 Pooling and filtering can qualitatively affect inferences from microbial diversity analyses (Ib)
The effect of soil treatment on aphid microbiomes was investigated using single and pooled aphid data sets separately in Buchnera-filtered and/or prevalence-based filtered data sets. Most statistical analyses resulted in similar outcomes. Differences in qualitative results based on the chosen analysis approach were observed in bacterial evenness, bacterial community composition and fungal species richness. While Buchnera-filtering affected bacterial evenness results, prevalence-based filtering affected bacterial evenness and fungal species richness results, and pooling affected bacterial community composition results and fungal species richness (Table 2). In bacterial community composition results, pooled aphid samples always showed soil treatment to be a significant factor explaining the variance within the data sets, while in single aphids, the factor soil was often not significant based on PERMANOVA results. On the contrary, the R2 values for soil microbiome being a significant factor explaining the fungal community composition were higher in single than in pooled aphids (Supporting Information S1: Table 2).
| With Buchnera, unfiltered | No Buchnera, unfiltered | With Buchnera, filtered | No Buchnera, filtered | Consistent in all analyses? | ||
|---|---|---|---|---|---|---|
| Bacteria | Species richness | Yes | Yes | Yes | Yes | Yes |
| Evenness | Yes | Yes | Yes | Yes | Yes | |
| Shannon | Yes | Yes | Yes | No | No | |
| Abundance | Yes | Yes | Yes | Yes | Yes | |
| Bray−Curtisb | No | No | No | No | No | |
| Bray−Curtisc | ||||||
| Clayey versus mixed | No | No | No | No | No | |
| Clayey versus sandy | Yes | No | Yes | Yes | No | |
| Mixed versus sandy | Yes | Yesa | No | No | No | |
| Weighted UniFracb | No | No | No | No | No | |
| Weighted UniFracc | ||||||
| Clayey versus mixed | No | Yes | Yes | Yes | No | |
| Clayey versus sandy | No | No | Yes | Yes | No | |
| Mixed versus sandy | No | No | Yesa | Yesa | No | |
| Fungi | Species richness | Yes | - | No | - | No |
| Evenness | Yes | - | Yes | - | yes | |
| Shannon | Yes | - | Yes | - | Yes | |
| Abundance | Yes | - | Yes | - | Yes | |
| Bray−Curtisb | Yesa | - | Yesa | - | Yes | |
| Bray−Curtisc | ||||||
| Clayey versus mixed | Yesa | - | Yesa | - | Yes | |
| Clayey versus sandy | No | - | No | - | No | |
| Mixed versus sandy | Yesa | Yesa | Yes | |||
| Weighted UniFracb | Yesa | - | Yesa | - | Yes | |
| Weighted UniFracc | ||||||
| Clayey versus mixed | Yesa | - | Yesa | - | Yes | |
| Clayey versus sandy | No | - | No | - | No | |
| Mixed versus sandy | Yesa | - | Yesa | - | Yes |
- Note: Diversity indices were modelled as a function of soil treatments, then, the results from single and pooled aphid data sets were compared. All analyses were performed with an unfiltered data set and a filtered data set using a prevalence-based 16% threshold (‘filtered’); for bacteria, analyses were again repeated using data sets with and without primary endosymbiont Buchnera aphidicola. Yes (green): same results were obtained for single and pooled data sets; no (red): results differed between single and pooled data sets; aR2-value is higher in single aphids; bPERMANOVA of distance matrix for factor ‘soil treatment’; cpairwise PERMANOVA for factor ‘soil treatment’. For detailed statistical results, see Supporting Information S1: Table 3.
3.3 Buchnera in T. annulatus supports the cospeciation hypothesis (II)
We found a total of 19 ASVs assigned to Buchnera in the full data set, with 17 ASVs found in pooled, five in single and three in both pooled and single aphid samples. Only one ASV was steadily displaying relative abundances >0.5% and was prevalent in all samples and thus regarded as representative. The mean relative abundance of Buchnera ASV was significantly higher in pooled than in single aphids (77.7 vs. 65.5%, t-test: p < 0.001). B. aphidicola of T. annulatus clusters within sequences derived from primary endosymbionts found in other Tuberculatus species and species within the Myzocallidini (Figure 2). The current analysis places the T. annulatus endosymbiont as the sister taxon to all other non-European aphid samples of the genus Tuberculatus in the data set (T. higuchii, T. capitatus, T. querciformosanus), but the phylogram topology is only weakly supported based on bootstrap values. Seven of the nonrepresentative ASVs do not follow cospeciation topology, of which only two ASVs (found in two and five samples) were found in >1 sample.
3.4 Secondary endosymbiont prevalence and abundance can be biased by pooling (III)
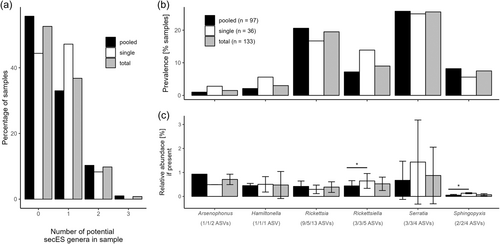
Around 44% and 56% of single and pooled aphid samples did not contain any known secES, respectively (Figure 3a). While >36% of all samples (pooled and single combined) included at least one, around 10% included two, and only one sample of pooled aphids contained three potential secES genera. Serratia and Rickettsia were the most prevalent genera (Figure 3b), but—if present—Serratia, Arsenophonus (in pooled aphid samples) and Rickettsiella (in single aphids) were the genera displaying the highest relative abundance (Figure 3c). Rickettsiella and Sphingopyxis showed significantly higher relative abundance in single compared to pooled aphid samples (Figure 3c). The relative abundance of secES exceeded 1% only in Serratia in seven samples (five in pooled, two in single aphid samples).
4 DISCUSSION
Microbiome research is constantly developing due to methodological advances. Despite the increasing number of available biostatistical tools for data analyses, the characteristics of a given data set need to be considered to choose an appropriate experimental, methodological and statistical design (Berg et al., 2020). Physical, chemical and biological differences between environmental samples (e.g., soil, roots, leaves, herbivores) often require different sample handling regarding both laboratory protocols and statistical analyses. Aphid microbiomes however display a very specific microbial community composition compared to other microbiomes like soil, rhizosphere or mammalian gut microbiomes. They are heavily skewed towards the primary endosymbiont B. aphidicola while containing a dynamic, but relatively low-abundant proportion of transient microbes. We hereby highlight these peculiarities and potential pitfalls in aphid microbiome analyses to ease future data handling and interpretation.
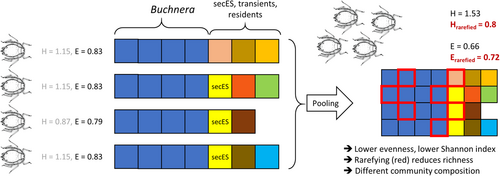
Pooling and in silico filtering often quantitatively affected microbial diversity indices in aphid microbiome analyses. Pooling to obtain sufficient amounts of DNA appeared to be decisive for sufficient DNA yield especially when amplifying fungal reads using PCR. Counterintuitively, bacterial alpha diversity indices were largely higher in single than in pooled aphid samples. This effect can be explained by the specific microbial community structure in aphids: one single Buchnera ASV dominates all samples and the remaining bacterial community is highly variable. When pooling aphids, the relative abundance of a taxon only found in part of the aphid is more likely to be below the detection threshold or to be lost in silico due to rarefying or filtering. This would result in a seemingly lower bacterial richness, evenness and Shannon diversity (Figure 4). Other methodological decisions like aphid surface sterilization, washing steps during aphid DNA extraction or using a 97% OTU threshold, may lead to underestimating the proportion of transient or resident ectosymbionts in aphids under natural conditions.
The discussion about how to analyse amplicon data sets is an ongoing topic in microbiome research (Boshuizen & te Beest, 2023). This includes the choice for the amplified genetic region (Johnson et al., 2019) as well as basic preprocessing steps like the usage of OTUs versus ASVs (Callahan et al., 2017; Caruso et al., 2019; Chiarello et al., 2022; Nearing et al., 2018), rarefying (Boshuizen & te Beest, 2023; McMurdie & Holmes, 2014) or filtering of spurious taxa (Cao et al., 2021; Nearing et al., 2022; Reitmeier et al., 2021; Schloss, 2020). In contrast to quantitative results, pooling of aphids and in silico filtering led to qualitatively similar results. Using T. annulatus, we observed differences in specific microbial diversity indices, while former work (Jousselin et al., 2016) reported no effect of pooling on bacterial microbiomes in the aphid genus Cinara. In contrast to T. annulatus, Cinara aphids are comparably big, rich in endosymbiotic bacteria, densely haired and adapted to conifers (Dransfield & Brightwell, 1999). For specific research questions, the potential effect of pooling aphids on microbial species richness, evenness and community composition may need to be considered.
Phylogenetic analyses of 16SrRNA genes placed the representative sequence of the primary endosymbiont B. aphidicola in T. annulatus clearly within other Tuberculatus species, supporting the cospeciation concept in oak aphids. On the other hand, we observed several ASVs that would not support cospeciation but appear in low prevalence and could be thus interpreted as sequencing errors or read joining errors. Therefore, using endosymbiont ASVs for phylogenetic analyses requires the sequence being checked to be representative for the aphid species beforehand. Representativeness does not necessarily lead to one specific ASV, since single aphids can also harbour two genotypes of one endosymbiont species (Guyomar et al., 2018). While the hereby used ASV gives first evidence, using the complete Buchnera 16SrRNA gene fragment would increase confidence values for the statement of phylogenetic congruence within Tuberculatus taxonomy. When sequencing 18SrRNA of several aphid species, Coeur d'Acier et al. (2014) found a comparable high intraspecific genetic variance in T. annulatus, indicating yet undescribed cryptic species or subspecies. Combining host genomic data with endosymbiont genomes may provide the necessary resolution to distinguish different aphid lineages within T. annulatus.
Former research indicates secES ratios in aphid populations to be fixed in aphids for a given species, even when reared under stable lab conditions (Jousselin et al., 2016), and secES are steadily represented in T. annulatus. Hamiltonella was reported in the genus Tuberculatus before (Henry et al., 2015), but we hereby report for the first time the genera Arsenophonus, Rickettsia, Rickettsiella, Serratia and Sphingopyxis in T. annulatus. Hamiltonella mediates protection against parasitoids to aphids, while Rickettsia and Rickettsiella can mediate protection against fungal pathogens (Zytynska & Weisser, 2016). Rickettsiella was further reported to lead to aphid colour changes (Zytynska & Weisser, 2016). Serratia is more abundant in oligophagous than in polyphagous aphid species (Henry et al., 2015), beneficial to the host under heat stress, functionally replacing or supplementing Buchnera, and the most frequent secES in aphids (Zytynska & Weisser, 2016). Sphingopyxis is mostly found in tree-adapted aphid species (McLean et al., 2019), as is the case for T. annulatus. At this point, an ectosymbiotic lifestyle and horizontal acquisition by the aphid cannot be excluded for the genus Sphingopyxis.
Since our results are yet only based on sequence data, the endosymbiotic lifestyle of the detected taxa still requires further confirmation by using other methods like FISH-CLSM. Noteworthy, both the prevalence and the mean relative abundance of the newly reported secES were low, meaning that using an in silico-filtered data set would have resulted in the conclusion of T. annulatus to be secES-free. Under field conditions, one to two secES can usually be detected in aphids, rarely up to four secES in generalist aphid species (Zytynska & Weisser, 2016). Multispecies secES infections increase both the protective as well as the detrimental effects of the endosymbionts (Zytynska & Weisser, 2016). We only found triple infections with secES in pooled aphid samples, while in single aphids we rarely found double infections. Therefore, our data suggests the maximum number of bacterial secES species in T. annulatus to be two under lab-rearing conditions. While secES prevalence was comparable in pooled and single aphid samples, their relative abundance tends to be higher in single aphid samples probably due to ‘dilution’ with nonhousing aphid DNA in pooled samples. We conclude that pooling leads to overestimating the number of double infections with secES, partially overestimating secES prevalence while underestimating their abundance in single aphid individuals.
5 CONCLUSIONS
To summarize, aphid microbiome research needs to consider the specific peculiarities of the microbial community composition in aphids in their experimental setup, data processing and when choosing statistical analyses. For phylogenetic analyses of Buchnera using ASVs, the obtained sequences need to be checked for representativeness. Sufficient bacterial DNA for amplicon sequencing can be obtained when extracting single aphid individuals, but fungal DNA yield and consequently PCR success is low. Comparative analyses of secES prevalence and abundance between aphid species should be based on single aphid sequencing, or otherwise only discussed with caution.
AUTHOR CONTRIBUTIONS
Adrian Wolfgang analysed the data and generated the visualizations. Adrian Wolfgang and Ahmed Abdelfattah wrote the manuscript. All authors designed the study and contributed to the final version of the manuscript.
ACKNOWLEDGEMENTS
The authors would like to thank Daniela Amhofer (Graz), Anaís Carpelan, Laura van Dijk (Stockholm University), Nora Temme and Ralf Tilcher (Einbeck). This work was funded equally by the European Union's Horizon 2020 under the ‘Nurturing excellence by means of cross-border and cross-sector mobility’ program for MSCA-IF-2018-Individual Fellowships, grant agreement 844114, and ‘BIOINSECTICIDES’ research project (F42422) at Graz University of Technology. Open Access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The authors confirm that they have adhered to the ethical policies of the journal.
Open Research
DATA AVAILABILITY STATEMENT
The data set supporting the conclusions of this article is available in the European Nucleotide Archive (ENA) repository, accession number PRJEB50358.




