Aegerolysins: Structure, function, and putative biological role
Abstract
Aegerolysins, discovered in fungi, bacteria and plants, are highly similar proteins with interesting biological properties. Certain aegerolysins possess antitumoral, antiproliferative, and antibacterial activities. Further possible medicinal applications include their use in the prevention of atherosclerosis, or as vaccines. Additional biotechnological value of fungal aegerolysins lies in their involvement in development, which could improve cultivation of commercially important edible mushrooms. Besides, new insights on microheterogeneity of raft-like membrane domains could be gained by using aegerolysins as specific markers in cell and molecular biology. Although the exact function of aegerolysins in their producing organisms remains to be explained, they are biochemically well characterized all-β structured proteins sharing the following common features: low isoelectric points, similar molecular weights (15–17 kDa), and stability in a wide pH range.
Introduction
In 2002, a new aegerolysin protein family (PF06355; IPR 009413) was defined, comprised of a few isolated and sequenced proteins, and several genomic transcripts and sequences predicted from expressed sequence tags. The first aegerolysin-like protein to be isolated1 and sequenced2 was Asp-hemolysin, a lethal and cardiotoxic component of the pathogenic fungus Aspergillus fumigatus. This opportunistic pathogen causes a wide spectrum of serious diseases including invasive aspergillosis. The pathogeneicity of A. fumigatus is a consequence of action of a series of toxic substances (cell wall components, allergens, pigments, adhesins, enzymes and toxins) which seem to act in an additive and/or synergic way on the cells.3 Asp-hemolysin was proposed to contribute to A. fumigatus pathogenesis.3 After its purification in the early seventies, a lot of work has been done to determine its structure and biological effects.
Subsequently, with the beginning of the genomics era, several other similar proteins were discovered. The first protein showing considerable amino acid sequence identity (43%) with Asp-hemolysin was the transcript of the Aa-Pri1 gene from the edible black poplar mushroom Agrocybe aegerita,4 which was soon followed by two putative hemolysin-like proteins from the bacterium Clostridium bifermentans.5 During the last few years, several other proteins or cDNA transcripts with highly identical sequences have been discovered6-11 and assigned to aegerolysins (Supporting Information). The isolated proteins and their recombinant forms were found to be acidic, to have a molecular weight from 15 to 17 kDa, and to share some important biological properties like membrane activity and a putative role in development.
The aim of this review is to summarize the current knowledge on this novel protein family, and to discuss the structure, function and possible biological role of aegerolysins.
Purification of Native and Recombinant Aegerolysin-Like Proteins
Much effort was invested in purification of the first aegerolysin-like protein, Asp-hemolysin from A. fumigatus. As this pathogenic mould produces various putative virulence factors, the purification was accomplished by monitoring the hemolytic activity during the isolation procedure, and separating the hemolytic compound from other toxic (e.g., dermonecrotic, neurotoxic, and nephrotoxic) molecules. Indications of a proteinaceous nature of the A. fumigatus hemolytic component can already be found in a very early report,12 which describes the loss of half of its toxicity after heating. This observation was later corroborated by other authors who had tried to purify the hemolytic compound(s) from the centrifuged mycelial homogenate.13, 14 The purification procedure was further improved by precipitation of the hemolytic compound with acetone and ammonium sulfate,14, 15 and column electrophoresis and ion-exchange chromatography on DEAE-cellulose.16 The protein was finally purified to homogeneity from the Fresenius-Muramatsu A. fumigatus strain, isolated from the lung of an infected penguin.1 The purification included precipitation with ammonium sulfate, anionic column chromatography and additional gel filtrations. This basic scheme was later used for purification of all other aegerolysins.7, 9, 17 In 2006, Ngai and Ng have simplified the purification procedure by gel filtration of crude fungal extract on FPLC, followed by ion-exchange chromatography and second gel filtration.
There are some peculiarities concerning the isolation of fungal aegerolysins. Ostreolysin and pleurotolysin A, for instance, can be isolated in 10-fold higher yields from freshly collected oyster mushrooms (Pleurotus ostreatus) than from frozen ones9(Berne, personal communication). Optimal isolation is achieved from primordia and young fruiting bodies of P. ostreatus, which coincides with the observation that the mushroom contains the highest concentration of hemolysin during this developmental stage.7, 18
Several expression constructs were designed to produce recombinant aegerolysins. Recombinant Asp-hemolysin was expressed in Escherichia coli as a fusion protein with maltose-binding protein (MBP) allowing a rapid, one-step purification on amylose resin.19 The purified protein exhibited the same immunoreactivity with anti-Asp-hemolysin antibodies as native Asp-hemolysin, and was able to bind the oxidized form of low-density plasma lipoprotein (ox-LDL), but did not show any hemolytic activity observed with the native protein. Efficient expression of Clostridium bifermentans cbm17.1 and cbm17.2 genes was achieved by introducing them as His6-tagged proteins in E. coli and Bacillus thuringiensis.5 However, when recombinant proteins Cbm17.1 and Cbm17.2 were assayed for biological activity, no toxicity to mosquito larvae was observed and no hemolytic action recorded.20 The His6-tagged PA0122 protein from Pseudomonas aeruginosa was also successfully expressed in E. coli.11 Purified recombinant PA0122 (rPA0122) protein was used for immunization and binding assays. It specifically bound to ox-LDL and lysophosphatidylcholine with an affinity equivalent to recombinant Asp-hemolysin, but similarly, did not exhibit any hemolytic activity.
In contrast to other aegerolysins, pleurotolysin is a two-component sphingomyelin-specific cytolysin.9 Recombinant pleurotolysin A (rPlyA) was expressed in E. coli as a soluble protein with the same molecular mass (17 kDa) and identical N-terminal amino acid sequence as natural PlyA. Recombinant pleurotolysin B (rPlyB) was obtained as a functional 59-kDa protein by unfolding and refolding of the insoluble E. coli fraction. In addition, recombinant PlyA and Ply B were immunostained with specific antisera against PlyA and PlyB, respectively. As each component lacked hemolytic activity in the absence of its counterpart, they were combined to study pleurotolysin pore-forming properties.21
Characterization of Aegerolysin-Like Proteins
The aegerolysin protein domain
In the Pfam protein database22 there are currently 43 members with the aegerolysin protein domain (PF06355) from 21 different species. Our PSI-BLAST23, 24 search (July, 2008) of the nonredundant protein database at NCBI (http://www.ncbi.nlm.nih.gov/BLAST), using the Aa-Pri1 translated amino acid sequence (O42717) from A. aegerita as query confirmed the Pfam entries. Additional fungal and plant homologs were retrieved from the expressed sequence tags (EST) database at NCBI, and from the Fungal Genome Initiative (FGI) database, Broad Institute (http://www.broad.mit.edu/annotation/fungi/fgi/) using the same query and the tblastn algorithm. In constructing our aegerolysin protein alignment (see Fig. 1), highly similar strain-specific sequences (e.g., 98% redundancy in Pseudomonas aeruginosa), and sequence fragments (e.g., in Bacillus thuringiensis) were omitted. Utilizing the consensus sequence, generated by Jalview,28 aegerolysin secondary structure was predicted by Jpred3,29 hyprospII,30 and PredictProtein.31 All three prediction tools generated almost identical results, proposing aegerolysins as all-beta proteins. This structural design was confirmed experimentally in the case of ostreolysin. CD spectroscopy revealed that this aegerolysin-like protein at pH 7.8 has 66% β-structure (6% β-turn and 60% β-sheet), 10% α-helix and the rest aperiodic secondary structure.32 These data agree with FTIR spectroscopy results.33

Protein alignment of the aegerolysin family members. Sequences were retrieved from Entrez (http://www.ncbi.nlm.nih.gov/sites/gquery) and manually edited to include only the aegerolysin domain for an optimal alignment generated by M-Coffee.25, 26 The alignment output was edited using GeneDoc27 conserved mode of shading, showing percentage of conservation (60%, light gray; 80%, grey; 100%, dark grey) within a column in the alignment. Consensus (boxed in red) generated by Jalview28 was used to predicit aegerolysin secondary structure. The distribution of secondary structure elements is presented bellow the protein alignment (green box-extended strand, grey line - random coil). The accession numbers of the proteins in Figure 1 are as follows: aegerolysin (gi 24636240), ostreolysin (gi 60461919), pleurotolysin_A (gi 54312022), T_versicolor (gi 90639437), H_annosum (gi 186897694), Asp-hemolysin (gi 70985747), A_clavatus (gi 121709507), A_nidulans (gi 67522192), A_niger_1 (gi 145230219), A_niger_2 (gi 145256342), A_oryzae_1 (gi 169777319), A_oryzae_2 (gi 169772307), A_oryzae_3 (gi 169785219), A_terreus (gi 115390458), N_fischeri (gi 119487614), B_cinerea (gi 116087898), C_immitis (gi 119182290), P_brasiliensis (gi 46346063), N_crassa (gi 85079504), B_vulgaris (gi 26112720), L_multiflorum (gi 46507636), R_australe (gi 158524422), P_aeruginosa_1 (gi 152985646), P_aeruginosa_2 (gi 15595320), C_bifermentans_1 (gi 2292821), C_bifermentans_2 (gi 2292820), B_thuringiensis_1 (gi 14571542), B_thuringiensis_2 (gi 49175503).
Aegerolysin protein domain, derived from the multiple sequence alignment and hidden Markov models (HMMs), is rich in aromatic (around 10%) and charged (20–24%) amino acid residues (pepstats predicition).34 Basidiomycete and Aspergillus aegerolysins contain a cluster of negatively charged amino acids (see Fig. 2). This segment, homologous to the ApoB-binding site of human low-density-lipoprotein receptor,35 was demonstrated to bind ox-LDL, and lysophosphatidycholine as a typical lipid moiety of ox-LDL.11, 36, 37 Furthermore, as proposed by Sakurai et al.,21 this negatively charged region may be involved in the recognition of choline moiety of sphingomyelin. Another feature of aegerolysin protein domain is conservation of cysteine and certain trypthophan residues (see Fig. 2) implying their possible structural and/or functional roles. Intact cysteine residues have been proven indispensable for hemolytic activity of ostreolysin7 and Asp-hemolysin.38 Tryptophan-rich regions have been reported in some other pore-forming proteins as responsible for the initial attachment to the membrane.39, 40 Moreover, combined aromatic residues and a cysteine residue have been suggested to be involved in sterol binding of cholesterol-dependent bacterial pore-forming toxins.41

Graphical display of aegerolysin protein domain. Shematic representation of aegerolysin domain is drawn (http://pfam.sanger.ac.uk/generateGraphic) and numbered according to Asp-hemolysin as most extensively studied aegerolysin-like protein. Red arrows indicate strictly conserved residues (Gly-75, Trp-94 and aromatic amino acid residue at position 114). Red diamonds above aegerolysin domain (blue) represent a cluster of negatively charged amino acid residues (at positions 39, 41, 43, 45, 49, and 50). Conserved cysteine residues (at positions 67 and 96) are depicted as green circles bellow protein domain. Tryptophans at positions 10, 33, 94, 98 are also conserved and marked with the black line bellow the domain.
Taxonomic distribution of aegerolysin-like proteins
Aegerolysin-like proteins appear in eukaryotic and bacterial taxa (Fig. 3). Most species (18) with genes encoding aegerolysins are found within Fungi, and belong to two sister groups, the Ascomycetes and the Basidiomycetes. In the former, the Aspergillus genus has the highest number od species (8) with aegerolysin genes. All fungal species with aegerolysin genes are either filamentous or dimorphic. Interestingly, the only other eukaryotic group with genes encoding aegerolysin-like proteins is Plantae. Blast searches revealed two eudicotyledonous and one monocotyledonous species to contain aegerolysin ESTs. Aegerolysin-like proteins also appear in two different taxonomic groups in Bacteria; the Firmicutes and the gamma-proteobacteria. To further diversify the distribution of aegerolysin-like proteins, there is also one viral representative, the Trichoplusia ni ascovirus 2c.
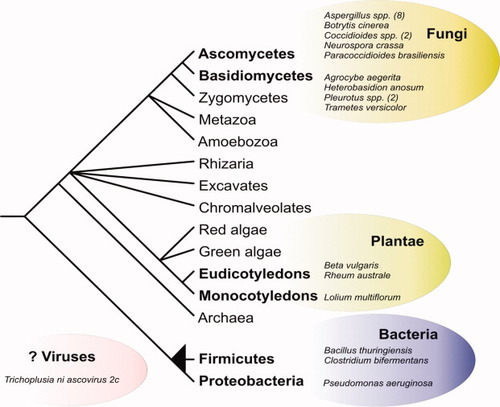
Taxonomic distribution of aegerolysin-like proteins. Schematic representation of eukaryote and prokaryote evolution where species or genera (number of species shown in parenthesis) with aegerolysin genes are shown in bubbles. The position of Viruses is uncertain. Drawn after Hedges,42 Abby and Daubin,43 and Keeling et al.44 [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The evolutionary relationships between the relatively few members of the aegerolysin protein family are difficult to establish and the phylogenetic tree is left unresolved (tree not shown). Well-supported phylogenetic relationships can be discerned only among the smaller taxa, for example, in Basidiomycetes, or the numerous strain-specific homologs in Bacillus thuringiensis. The latter is perhaps a case of microheterogeneity, where numerous aegerolysin-like homologs constitute a set of structurally similar, yet functionally diverse proteins.45
As to the origin of the aegerolysin-encoding gene, we can hypothesize that it first appeared in Fungi, in the last common ancestor of Ascomycetes and Basidiomycetes, where it is present in the greatest number of species. This assumption could be supported by the putative biological role of these proteins in fruiting body development, signaling, and their role as potential virulence factors in pathogenic species. There were only three species of the numerous sequenced bacterial genomes found to contain aegerolysin-encoding genes, belonging to two divergent taxa. A readily available explanation for bacterial aegerolysin family members is horizontal gene transfer (HGT) from fungi. Fluorescent pseudomonads were found to be closely associated with the cultivated mushroom Pleurotus ostreatus,46 which is a rich source of aegerolysin-like proteins.7, 9 The incorporation of aegerolysin homologs into bacterial genomes could be beneficial in the same respects as their putative biological roles are beneficial to fungi.
More puzzling is the appearance of aegerolysins in plants, again found in only three species belonging to monocots and dicots, in the EST database. This might also be a case of HGT, although reports of HGT to plant nuclear DNA are rare.47 HGT could have had occurred either from fungi or bacteria, which both form an intimate relationship with the host organism as plant pathogens or symbionts. Such close ecological associations could provide opportunities for gene transfer events.48 It could, however, also be a result of plant samples contaminated with mould (e.g., Lolium multiflorum leaf samples infected with powdery mildew [Ikeda, S., 2004 (unpublished), EST sequence database at NCBI)].
Physical and biochemical characterization of aegerolysin-like proteins
Proteins from the aegerolysin family share several common features. They all have very low isoelectric points, similar molecular weights, and are heat-labile (see Table I). Furthermore, they seem to be stable in a wide range of pH.1, 10, 32 In contrast to most of the known aegerolysins, that exert their biological properties (mainly the membrane lytic properties) in their monomeric forms,1, 7, 10, 17 pleurotolysin A isolated from the mushroom Pleurotus ostreatus acts as a bi-component cytolysin in concert with a 59 kDa pleurotolysin B. When coupled, these two proteins assemble into a ring-shaped transmembrane pore and are named pleurotolysin.9, 21
| Species | Protein | Prot param (ExPASy Tools) | Exp. Data | Reference | |||
|---|---|---|---|---|---|---|---|
| Amino acids | MW (Da) | pI | MW (Da) | pI | |||
| Agrocybe aegerita | Aegerolysin Aa-Pri1 precursor | 145 | 16,104 | 5.65 | 16,093 | 5.7 | 4 |
| Aegerolysin | 10 | 14,898 | 4.9 | 50 | |||
| cDNA clone Agf19 | 145 | 16,087 | 6.10 | Im, Lee and Shim, 2006 (unpublished) | |||
| Heterobasidion anosum | cDNA clone 18A12 (2a) | 130 | 14,236 | 6.07 | 51 | ||
| Pleurotus eryngii | Eryngeolysin | 40 | 17,000 | 10 | |||
| Pleurotus ostreatus | Ostreolysin | 137 | 14,855 | 4.82 | 32 | ||
| Ostreolysin | 50 | 17,000 | 5.0 | 7 | |||
| Ostreolysin | 14,975 | 52 | |||||
| Pleurotoysin | 12,600a | 6.4 | 17 | ||||
| Pleurotolysin A | 138 | 15,136 | 5.87 | 21 | |||
| Pleurotolysin A | 17,000 | 9 | |||||
| Trametes versicolor | cDNA clone TverSEQ10942 | 174 | 19,205 | 5.05 | Tsang, Storms and Butler, 2006 (Unpublished) | ||
| Aspergillus fumigatus | Asp-hemolysin precursor | 139 | 15,199 | 5.29 | 53 | ||
| Asp-hemolysin | 30,000b | 4.0 | 1 | ||||
| Asp-hemolysin | 131 | 14,275 | 5.24 | 2 | |||
| Asp hemolysin-like protein | 139 | 15,820 | 4.54 | 53 | |||
| Aspergillus clavatus | Hypothetical protein ACLA_066510 | 139 | 15,644 | 4.57 | Nierman, 2006 (Unpublished) | ||
| Aspegillus flavus | Conserved hypothetical protein AFL2G_03935 | 139 | 15,571 | 4.61 | FGI Broad Institute | ||
| Predicted protein AFL2G_08431 | 104 | 11,365 | 4.55 | FGI Broad Institute | |||
| Aspergillus nidulans | Hypothetical protein AN1553.2 | 136 | 14,864 | 4.59 | Birren et al. 2003 (Unpublished) | ||
| Aspergillus niger | Hypothetical protein An01g09980 | 145 | 16,259 | 7.81 | 54 | ||
| Hypothetical protein An19g00210 | 142 | 16,256 | 5.20 | 54 | |||
| Aspergillus oryzae | Unnamed protein product AO090701000257 | 139 | 15,571 | 4.61 | 55 | ||
| Unnamed protein product AO090023000032 | 141 | 16,359 | 5.10 | 55 | |||
| Unnamed protein product AO090010000018 | 144 | 16,294 | 9.53 | 55 | |||
| Aspergillus terreus | Conserved hypothetical protein ATEG_03556 | 152 | 17,022 | 5.24 | Birren et al. 2005 (Unpublished) | ||
| Neosartorya fischeri | Asp hemolysin-like protein NFIA_030750 | 139 | 15,810 | 4.64 | Nierman et al. 2006 (Unpublished) | ||
| Botrytis cinerea | cDNA clone 3498_G06 | 144 | 15,795 | 5.47 | 56 | ||
| Coccidioides immitis | Hypothetical protein CIMG_06184 | 182 | 20,269 | 6.30 | Birren et al. 2005 (Unpublished) | ||
| Coccidioides posadasii | Hypothetical protein CPAG_03541.1 | 134 | 14,798 | 5.01 | FGI Broad Institute | ||
| Neurospora crassa | Hypothetical protein NCU03475 | 198 | 21,401 | 5.05 | 8 | ||
| Paracoccidioides brasiliensis | cDNA clone EST008190 | 127 | 14,078 | 3.88 | 57 | ||
| Beta vulgaris | cDNA clone 024-006-B13 | 135 | 14,875 | 6.06 | 58 | ||
| Lolium multiflorum | cDNA clone SL010G09-5 | 135 | 14,691 | 6.08 | Ikeda, 2004 (Unpublished) | ||
| Rheum australe | cDNA clone RaSFL 362 | 116 | 12,830 | 4.51 | Ghawana et al. 2007 (Unpublished) | ||
| Bacillus thuringiensis | Crystal protein ET80 | 132 | 14,839 | 6.02 | 59 | ||
| Cry34A-like protein | 97 | 10,614 | 5.86 | 60 | |||
| Clostridium bifermentans malaysia | Hemolysin-like protein Cbm 17.1 | 153 | 17,200 | 5.41 | 5 | ||
| Hemolysin-like protein Cbm 17.2 | 152 | 17,463 | 5.82 | 5 | |||
| Pseudomonas aeruginosa | Hypothetical protein PA0122 | 136 | 14,579 | 4.65 | 6, 11 | ||
| Aegerolysin superfamily | 121 | 12,713 | 8.36 | Dodson et al. 2007 (Unpublished) | |||
| Trichoplusia ni ascovirus 2c | Hypothetical protein TNAV2c_gp029 | 222 | 23,777 | 6.19 | 60 | ||
- a Lower MW presumably due to proteolytic degradation.
- b Probably protein aggregation.
Biological Activities of Aegerolysin-Like Proteins
Interaction with lipid membranes
Hemolytic activity
The first isolated aegerolysin, Asp-hemolysin, was proposed to form distinctive pores in erythrocyte membranes and to induce hemolysis by a colloid-osmotic mechanism. An early study using scanning electron microscopy showed that this hemolysin forms rings and patches at the surface of erythrocyte membranes.61 Aggregation of the protein at the membrane surface and inhibition of hemolysis using osmotic protectants of various molecular weights was later proven also for ostreolysin,33 pleurotolysin,9 and eryngeolysin.10 The hydrodynamic diameters of the resulting pores were estimated to be around 4 nm.9, 33 Pleurotolysin-induced lysis was reported to proceed (i) by the initial binding of the 17 kDa sphingomyelin-binding component, pleurotolysin A, to the membrane, (ii) subsequent addition of pleurotolysin B (59 kDa), and (iii) their association into a 700 kDa SDS-stable, ring-shaped structures with outer and inner diameters of 14 and 7 nm, respectively. Most efficient hemolysis occurred when the two components were mixed in a 3:1 molar ratio, implying that they are incorporated in the pore complex in the same molar ratio.9 All aegerolysins aforementioned were hemolytic in the nanomolar range.
The pH-optima for binding of aegerolysins to the erythrocyte membrane and its permeabilization do not overlap. Optimal binding is usually achieved at pH 5–7, as shown for Asp-hemolysin61 and ostreolysin.32 In contrast, the maximal hemolysis is observed at pH 7–8.17, 32 Aegerolysins however retain their membrane activity in a wide pH range from 3.5 to 10.5.1, 10, 32, 61 Binding of ostreolysin to the membrane correlates with an increase of intrinsic tryptophan fluorescence and the α-helical content of the protein, and with aggregation, suggesting that binding to the bilayers evokes further conformational transitions necessary for insertion and pore formation.32, 33
Asp-hemolysin lytic activity can be abolished by certain divalent ions, like Hg2+, Cu2+, Fe2+, and Pb2+, and later restored by the use of β-mercaptoethanol or cysteine.1 Similar inhibition of hemolytic activity by micromolar concentrations of Hg2+ and Fe2+ was also reported for ostreolysin32 and eryngeolysin,10 respectively. Early studies involving chemical modifications of Asp-hemolysin suggested that while cysteines and arginines were necessary for hemolytic activity,38, 62-64 histidine residues were not. The implication of intact cysteine residues in a hemolytic process was later proven also for ostreolysin.7, 32
Varying interaction with erythrocytes from different species is another interesting feature of assorted aegerolysins. For example, Asp-hemoylsin is more lytic to chicken and human erythrocytes than to those of rodents.1 Its activity against frog erythrocytes is 833-fold lower than against chicken erythrocytes.65 A slightly different pattern, showing greater sensitivity of mammalian erythrocytes in comparison to avian ones, was observed for eryngeolysin.10 The membrane activity of pleurotolysin was proposed to correlate with the sphingomyelin content of erythrocyte membranes, exhibiting the following preferences: sheep > ox > goat > man > cat > rabbit > swine > mouse/rat > horse > dog > guinea pig,17 or sheep > human > rabbit > dog > horse.9 In contrast, ostreolysin-induced lysis of sheep, bovine and human erythrocytes33 was similar, but much less potent against canine66 and rodent67 erythrocytes.
Permeabilization of lipid vesicles
The lytic process of many pore-forming proteins starts with the recognition of a distinct membrane component. The binding of eryngeolysin, for example, was found to be inhibited by millimolar concentrations of N-glycosylneuraminic acid.10 The aegerolysins' lytic activity, however, is not altered upon incubation with pure sphingo- or glycerophoypholipids, or cholesterol.33, 65 Similarly, liposomes composed of egg phosphatidylcholine in combination with various other lipids were unable to inhibit ostreolysin-induced hemolysis.33 In contrast, liposomes composed from sheep erythrocyte sphingomyelin and cholesterol in a 1:1 molar ratio successfully inhibited binding and lytic activity of pleurotolysin,17, 9 and ostreolysin.51 As these effects were absent when liposomes were composed of cholesterol in combination with other lipids, it was proposed that pleurotolysin is a specific sphingomyelin-binding protein.9, 17 However, recent work performed on ostreolysin suggests that the specific interaction of this protein (and probably other aegerolysin-like proteins) with sphingomyelin:cholesterol liposomes could be due to its binding to cholesterol-enriched raft-like membrane microdomains. These highly investigated membrane entities were reported to be involved in several important biological functions, such as exocytosis and endocytosis, signal transduction, pathogen entry, and attachment of various ligands,68-70 and to exist in a so-called liquid-ordered phase. Lipid domains in such a physical state are more resistant to solubilization by detergents than the ones in a liquid-disordered phase.71 Hence, lipid rafts are operationally often called detergent-resistant membranes (DRMs).68 Upon preincubation, ostreolysin was found in isolated DRMs of both sphingomyelin:cholesterol vesicles and Chinese Hamster Ovary cells.51 Further, permeabilization of sphingomyelin:cholesterol (1:1) vesicles by ostreolysin occurred only above 30 mol% cholesterol, the concentration at which this sterol induces the formation of a liquid-ordered phase.51, 72, 73 The interaction of ostreolysin with cholesterol-enriched membrane domains can be diminished or disrupted (i) by the addition of mono- and di-unsaturated phosphatidylcholine,51 (ii) by replacing cholesterol with other natural sterols or cholesterol derivatives,73 (iii) by the addition of micromolar concentrations of fatty acids and lysophospholipids,33, 74 and (iv) by the pre-treatment of cell membranes with a cholesterol scavenger, methyl-β-cyclodextrin.74 Ostreolysin did not affect electron paramagnetic resonance spectra of sphingomyelin:cholesterol (1:1) vesicles,51 which would indicate a recruitment of the lipids upon its binding to the membrane, a feature that was proven for other raft-binding cytolysins like the cholera toxin B subunit.75 It seems that this protein, rather than recognizing sphingomyelin, recognizes and binds to specific patterns formed by cholesterol molecules at the surface of biological membranes.
Cholesterol-dependence of ostreolysin resembles that of bacterial thiol-activated (cholesterol-dependent) cytolysins (CDCs), allowing us to conclude that this protein, and probably other aegerolysin members, could be functionally classified as CDCs.73 Ostreolysin cannot bind pure cholesterol,33 but this sterol seems to be crucial for its membrane activity, which is highly cooperative with respect to cholesterol membrane concentration.51, 73 Similarly, it was recently shown that the sole presence of cholesterol is not a prerequisite for the membrane activity of CDCs,41, 75-78 suggesting furthermore that the distribution of cholesterol molecules on the membrane, and the specific patterns that are subsequently formed, could be of crucial importance for the activity of this group of cytolysins. For example, a cholesterol-binding pore-forming Vibrio cholerae cytolysin, whose crystal structure strongly resembles those of CDCs,79 can bind to pure cholesterol and oligomerize with it in solution, but it prefers association with sphingolipid/cholesterol complexes rather than with individual lipid molecules.80
Finally, the discovery that some aegerolysins bind specifically, and with high affinity to the ox-LDL11, 81 has lead to a more detailed research of interaction between the two molecules. The direct binding of lysophosphatidycholine (which is the major component of ox-LDL) to some aegerolysins, Asp-hemolysin and PA0122, was confirmed by ion-exchange chromatography37 and by surface plasmon resonance experiments respectively.11 Although the membrane activity of ostreolysin was strongly inhibited by lysophospholipids and fatty acids,33, 74 the same binding specificity could not be proven. It is suggested rather that lysophospholipids influence the membrane activity of ostreolysin by altering the physical properties of the membrane, for example, by partitioning into liquid-ordered membrane domains.74
Binding to serum lipoproteins
The membrane activity of aegerolysin-like proteins can be inhibited by the addition of serum.33, 65, 82 Indeed, binding to blood plasma components is another distinctive feature of the most studied aegerolysin-like protein, Asp-hemolysin.83 This protein is similar (26.5% identity and 68.8% similarity over a 34 amino acid overlap) to a human LDL receptor precursor2 and interacts with several lipoprotein components of human plasma, especially ox-LDL.35, 36, 84 The binding to ox-LDL is concentration-dependent, and is reduced if positively charged amino acid residues of the hemolysin are blocked.85 Surface plasmon resonance experiments confirmed the specific binding of Asp-hemolysin to ox-LDL with a Kd of 0.63 μg/mL.81 The binding constants of a recombinant aegerolysin from Pseudomonas aeruginosa (rPA0122) to ox-LDL and lysophosphatidylcholine were very similar to those of Asp-hemolysin.11
Recently, a synthetic peptide (P21) derived from the positively charged region of Asp-hemolysin was found to inhibit the ox-LDL-induced macrophage proliferation.86 Further, using synthetic peptides of 4–29 amino acid residues, the specific ox-LDL-binding sequence (YKDG) was identified.87 The YKDG region is important for binding of these peptides to lysophosphatidylcholine, that seems to be responsible for most of the effects of ox-LDL, including induction of macrophage proliferation,88 which has an important role in the development and progression of atherosclerosis.
Cytotoxic activity
The toxicity of aegerolysins to experimental animals is most probably also a consequence of their cytolytic effects. So far, aegerolysins were found to be cytotoxic for different cell lines (Table II). Direct microscopic observation of certain tumoral cell lines exposed to ostreolysin showed typical signs of a colloid-osmotic hemolysis, like swelling, blebbing and degranulation of cells.33 Similar events were noted during real-time monitoring of ostreolysin-induced disruption of fluorescently stained human chondrocytes and osteoblasts using confocal microscopy.91 Cytotoxicity was found to be affected by the same factors as hemolysis, for example, by chemically blocking the amino- and thiol groups, or by the serum.51, 82 Furthermore, macrophages treated with Asp-hemolysin express mRNA transcripts for several cytokines like TNF-alpha, interleukin-1a and 1b.82 Asp-hemolysin-induced mRNA expression of cytokines such as TNF-alpha, IL-1b, IL-6, IL-8, and granulocyte-macrophage colony stimulating factor was also reported for endothelial cells.90 Finally, eryngeolysin possesses antibacterial activity towards Bacillus subtillis and Bacillus megaterium, and inhibits the mitogenic response of murine splenocytes.10
| Protein | Cell line | ED50 (μg/mL) | Time of exposure (h) | Reference |
|---|---|---|---|---|
| Asp-hemolysin | Leukocytes (human) | 500 | NA | 90 |
| Alveolar macrophages (guinea pig) | 60 | NA | 90 | |
| Peritoneal macrophages (mouse) | 25 | 1 | 84 | |
| Umbilical vein endothelial cells (human) | 100 | 1 | 91 | |
| Ostreolysin | HT 1080 (fibrosarcoma, human) | 10 | 2 | 33 |
| MCF 7 (mammalian tumour, human) | 10 | 2 | 33 | |
| Chinese Hamster Ovary cells | 1 | 0.25 | 52 | |
| Human articular chondrocytes | 1 | 1 | 92 | |
| Fibroblasts (lungs of Chinese hamster, V-79-379A | 1.3 | 1 | 93 | |
| Endothelial cells (human umbilical vein, HUVEC) | 2.2 | 1 | 93 | |
| Eryngeolysin | L1210 leukemia (human) | NA | 10 | 10 |
- NA, not available.
Toxicity for experimental animals
Toxic and lethal effects of aegerolysin-like proteins were most extensively studied in Asp-hemolysin from Aspergillus fumigatus. This opportunistic pathogen has been known for a long time to cause neurotoxic,12, 93 dermonecrotic,94, 95 nephrotoxic,94, 96 complement-inhibitory,97 and hemolytic effects.93, 94 The LD50 of Asp-hemolysin for mice and chicken was determined to be 750 and 350 μg/kg, respectively. This presumably secretory protein2, 98 was also detected in vivo during the experimental infection of mice by A. fumigatus spores.98 Histopathological findings in mice after the i.v. administration of Asp-hemolysin comprise perivascular lesions of various degrees in kidney, heart, liver and brain, increase of capillary permeability, contraction of the ileum and cytotoxicity, accompanied by the appropriate biochemical alteration of sera.89, 99
Although the edible oyster mushroom (Pleurotus ostreatus) is a widely cultivated species, sporadic local intoxications following its ingestion by humans and animals were also recorded. It was suggested that the toxicity is associated with thermolabile proteinaceous molecules.100 Bernheimer and Avigad17 did not observe any obvious signs of intoxication in mice after i.v. application of 320 hemolytic units of pleurotolysin. However, recent investigation of ostreolysin revealed cardiorespiratory and toxic effects in rodents, and their death after an i.v. administration with a LD50 of 1175 μg/kg.67 Cardiotoxic effects are probably due to ostreolysin-induced dose-dependent increase in aortic ring tension, and its cytotoxicity to endothelial cells resulting in hyperkalaemia.92 The histopathological effects of ostreolysin in mice were not studied yet. Pleurotus ostreatus water extract, however, was found to provoke effects similar to those induced by Asp-hemolysin: hemorrhages in the intestine, liver, lung and kidney, and degenerative changes in the liver, with a LD50 higher than 3000 μg/kg.100 Such an extract also induced dose-dependent contractions of nonsensitized guinea pig trachea smooth muscle.101
Role of aegerolysins in development
Experimental results imply that aegerolysins play an important role in the growth cycle of the organism that produces them. Fernandez-Espinar and Labarère4 determined that Aa-Pri1 mRNA expression coincided with the developmental stage of primordia, and proposed an involvement of the putative protein in the compaction of hyphae in primordia.4 This protein was later purified and named aegerolysin.7 A detailed monitoring of the occurrence of ostreolysin and aegerolysin in mycelia and fruiting bodies of respective mushrooms (P. ostreatus and A. aegerita), and its correlation with the developmental stages of the fungi was performed using immunocytochemical methods. Both proteins were identified in the rapidly growing primordia, in the basidia and basidiospores of maturing fruiting bodies, which might indicate their function in initiation of fructification and/or sporulation.18 Their occurrence in peripheral parts of the fruiting bodies and lamellae suggests, in particular, that they may participate in differentiation of hyphae and formation of basidia and basidiospores. The latter hypothesis is supported by the finding that some bacterial aegerolysin-like proteins are expressed during sporulation.5 It is interesting to note that ostreolysin can markedly induce fructification of P. ostreatus even when applied externally to the mycelium.102
Perspectives
Although their physiological role in the producing organisms is not yet well understood, the biological properties of aegerolysin-like proteins make them interesting from several points of view. Asp-hemolysin, for example, is a lethal toxin that constitutes a very important part of the toxic battery of Aspergillus fumigatus during infection. This fungus causes a wide range of serious diseases, especially highly lethal invasive aspergillosis that often occurs in immunocompromised individuals, such as leukemia, cancer or AIDS patients, and after bone marrow and organ transplantations.103 Animal models have shown that anti-Asp-hemolysin IgG can successfully protect mice from the infection propagated with spores,98 and indicate that, beside the classical treatment with amphotericine, the use of anti-Asp-hemolysin IgG could be one of the strategies to protect immunocompromised patients from infection propagated by A. fumigatus. Moreover, the specific binding of Asp-hemolysin to ox-LDL makes this protein, its nonhemolytic recombinant mutants or synthetic forms86 a possible tool for investigation of the pathophysiological significance of ox-LDL.81 Further study on the binding mechanism between the Asp-hemolysin-related peptide and ox-LDL may provide important information on the prevention and treatment of atherosclerosis.
Specific binding to distinct membrane domains seems to be another characteristic feature of aegerolysin-like proteins, and has been most extensively studied in ostreolysin.51, 73, 74 Ostreolysin, and probably also other aegerolysins, specifically bind to raft-like cholesterol-rich membrane domains, and thus might be interesting as new tools for investigating these cell membrane domains. Their fluorescently- or spin-labeled mutants devoid of lytic activity could be useful in structural and functional studies of biological membranes. Indeed, several cytolytic proteins that specifically recognize particular components enriched in lipid rafts have been recently proposed as markers of raft-like membrane domains. A truncated, nontoxic mutant of lysenin (isolated from the earthworm Eisenia foetida) is a specific sphingomyelin-binding protein,104 cholera toxin B subunit (CT-B) binds to ganglioside GM1,105 and a nontoxic derivative of perfringolysin O from Clostridium perfringens requires higher cholesterol content as a prerequisite for binding.106 In contrast to these cytolysins, ostreolysin was shown to specifically sense the combination of the two main lipid components of membrane rafts - cholesterol and sphingomyelin.51 Thus, this protein could be proposed as a useful external lipid raft marker, able to detect domains ascribed to cholesterol-sphingomyelin rafts. Ostreolysin does not colocalize with other raft-binding proteins like caveolin or the commercially available cholera-toxin B subunit.74 Hence, new insights on microheterogeneity of cholesterol-enriched membrane microdomains could be gained by using ostreolysin-like probes.
Last but not least, fungal aegerolysins are specifically expressed during the early phases of fructification.4, 7, 18 Stimulation of fruiting in mushrooms can be achieved by a variety of external stimuli, including substances of natural or synthetic origin, reduced temperature, light, and nutrient depletion.102 Better understanding of genes, pathways and cellular processes leading to the initiation and development of fruiting bodies in fungi may improve cultivation of edible mushrooms. In this regard, the ability of ostreolysin to enhance the fructification of oyster mushrooms, and possibly of other mushrooms when applied externally102 makes this protein and its genetically engineered recombinant forms interesting from the commercial and biotechnological points of view.




