The Phenix-AlphaFold webservice: Enabling AlphaFold predictions for use in Phenix
Review Editor: Nir Ben-Tal
Abstract
Advances in machine learning have enabled sufficiently accurate predictions of protein structure to be used in macromolecular structure determination with crystallography and cryo-electron microscopy data. The Phenix software suite has AlphaFold predictions integrated into an automated pipeline that can start with an amino acid sequence and data, and automatically perform model-building and refinement to return a protein model fitted into the data. Due to the steep technical requirements of running AlphaFold efficiently, we have implemented a Phenix-AlphaFold webservice that enables all Phenix users to run AlphaFold predictions remotely from the Phenix GUI starting with the official 1.21 release. This webservice will be improved based on how it is used by the research community and the future research directions for Phenix.
1 INTRODUCTION
With recent advances in machine learning, it is now possible to use an amino acid sequence to predict the three-dimensional structure of proteins with sufficient accuracy to be useful as hypotheses for the structure (Terwilliger, Liebschner, et al., 2023). Software, like AlphaFold (Jumper et al., 2021), has revolutionized the workflow of protein structure determination by predicting highly accurate starting models that can then be used for fitting into data from crystallography or from cryo-electron microscopy (Barbarin-Bocahu & Graille, 2022; McCoy et al., 2022; Terwilliger, Afonine, et al., 2023; Terwilliger et al., 2022).
Phenix (Liebschner et al., 2019), a widely-used software suite for macromolecular structure determination, recently integrated AlphaFold version 2.2.2 into its program for building and refining structures, phenix.predict_and_build (Terwilliger, Afonine, et al., 2023; Terwilliger et al., 2022). Given the sequence of the protein and the data, either from crystallography or from cryo-electron microscopy (cryo-EM), Phenix can automatically generate predictions with AlphaFold, use the highly accurate parts of the predictions for building the model, and iteratively improve the model against the data. As part of this process, the pipeline will also feedback the updated models as templates for subsequent AlphaFold predictions to generate structures that are closer matches to the experimental data. This feedback loop requires that new predictions be made based on the sequence and template for the experimental sample, which precludes the use of databases with pre-calculated predictions (Jumper et al., 2021; Varadi et al., 2022).
For the average Phenix user, this powerful process of automated structure determination is limited by the technical challenges of running AlphaFold. The documented way for running AlphaFold is on a Linux-based computing platform with a high-performance NVIDIA graphics processing unit (GPU). The longer the sequence, the more memory is required on the GPU and the longer the time required to make the prediction. There is a way to run AlphaFold in a Google Colaboratory notebook (Bisong & Google Colaboratory, 2019) via ColabFold (Mirdita et al., 2022), but that poses problems for automation and reproducibility since the cloud notebook software and hardware specifications are regularly updated by Google with no control by the user. That is, if a user wanted to reproduce an earlier job with the same software, it would not be possible because Google manages the virtual machine running the underlying software for ColabFold, and currently, there is no way to consistently run earlier versions of that software. Furthermore, in order for AlphaFold to make protein structure predictions, multiple databases, approximately 2.4 TB in total size, are required. These database include all the sequences and structures in the Protein Data Bank (Berman et al., 2003), the Uniclust30 (Mirdita et al., 2017), and UniRef90 (Suzek et al., 2015) clustering of the sequences in UniProt at 30% and 90% sequence identity, respectively, the MGnify sequence database (Richardson et al., 2023), and the BFD sequence database (Jumper et al., 2021; Steinegger et al., 2019; Steinegger & Söding, 2018). By requiring a GPU and such large databases, AlphaFold would limit the usage of the Phenix automated pipeline.
Moreover, based on the download statistics of the Phenix installers from the Phenix website over the past year, around 25% of the downloads are for Linux, and the remainder of downloads is about evenly split between macOS and Windows (about 37.5% each). While 25% of the Phenix user base has the matching operating system for running AlphaFold, the actual percentage will likely be lower due to the hardware requirements. For example, it is unlikely that there is an affordable Linux laptop that would have multiple terabytes of disk storage and a high-end GPU. Furthermore, the Phenix user base runs the gamut from academic research groups with little computing support to large companies with dedicated staff for managing hardware and software. To support our user base, we want a solution that can make the phenix.predict_and_build tool generally available to all.
This computing problem has been recognized by others working on software and methods development in structural biology and many of the solutions like CCP4-cloud (Krissinel et al., 2018, 2022), HADDOCK (Dominguez et al., 2003; van Zundert et al., 2016), and Protein Data Bank (PDB)-REDO (Joosten et al., 2014; van Beusekom et al., 2018) use remote computing resources for their users. Users generally register for an account and upload their data to the server. CCP4-cloud also has modes where some software is run locally and some remotely, or entirely locally.
2 RESULTS
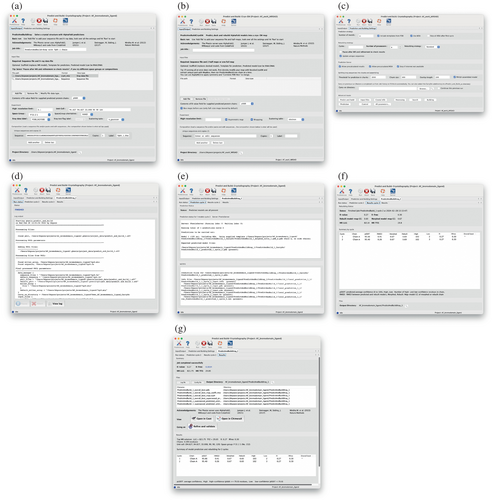
To make the process of running the Phenix automated pipelines that use AlphaFold generally available to all researchers implemented a Phenix-AlphaFold webservice at https://prediction.phenix-online.org that will accept automated pipeline jobs and run the AlphaFold portion of the job. The other parts of the process, like model building and refinement, are still run locally on the user's computer. This interface is available in the Phenix GUI starting with the 1.21 official release. Figure 1 shows the phenix.predict_and_build interface in the Phenix GUI (Echols et al., 2012), Figure 1a for crystallography data, Figure 1b for cryo-EM data, and Figure 1c for the common options for both data types. Users interact with these GUIs just like they do with other Phenix programs. After submitting a job with the sequence and required data, the GUI manages the communication between the user's computer and the Phenix-AlphaFold webservice by sending the sequence to the server and for retrieving the predicted model and associated prediction confidence statistics.
To provide flexibility for the user and resilience even in unstable network situations, we do not want the webservice to require a constant connection between the server and the Phenix GUI. To achieve this, we implemented the webservice with an Application Programming Interface (API) based on Representational State Transfer (REST). Because RESTful interfaces are “stateless,” a stable network connection to the webservice is not needed to manage the current status or “state” of the job. Each request from the Phenix GUI is independent and jobs are identified by a unique id. The most common implementation of a REST API was chosen, which uses standard Hypertext Transfer Protocol (HTTP) operations and the JavaScript Object Notation (JSON) data format. The webservice is available at https://prediction.phenix-online.org and Table 1 lists some of the basic commands that are used for starting jobs, querying the status of a job, getting the results from a job, and querying the status of the server. By using standard internet protocols and formats, the Phenix-AlphaFold webservice is easily accessible from any computer connected to the internet.
| URL command | JSON payload keys | JSON response keys |
|---|---|---|
| start_job | token program args calling_ip requires_gpu |
job_id status stdout stderr result job_dir |
| job_status | token job_id |
job_id status stdout stderr result job_dir |
| get_job_result | token job_id |
job_id status result stdout stderr |
| server_status | token | waiting_jobs running_jobs n_available_zmq_workers n_available_async_workers n_running_zmq_jobs n_queued_zmq_jobs n_complete_jobs wait_time_sec running_time_sec |
- Note: The payload keys are text arguments that are provided to the server. The “token” is for authorization and is needed to interact with the server. Currently, only jobs from the Phenix GUI are authorized. The “job_id” is a unique identifier for each job that is submitted. It is used for querying the status of the job as well as getting the result once it is finished. The “program” defines the program that is to be run on the server. We are currently limiting this argument to programs related to phenix.predict_and_build. The “args” parameter defines the parameters that are passed to the program. Generally, these would be the same arguments that would be passed to the command-line program. The “calling_ip” is the IP address of the computer making the request and “requires_gpu” specifies if a GPU is required to run the program. The response keys are returned by the server and parsed by the Phenix GUI. The important ones are “status,” “stdout,” “stderr,” and “result.” They let the Phenix GUI know about the job's status, the log output, any error output, and the output of the job, respectively.
To interact with the Phenix-AlphaFold webservice, the Phenix GUI uses the requests (Requests Developers, n.d.) Python package for constructing the commands to send to the webservice and for parsing the output that is received. When the Phenix GUI sends commands to the webservice, the GUI converts Python data structures into text stored in Python dictionaries. These dictionaries are converted into text formatted as JSON and sent to the server. The response from the server is also in JSON format which is converted into Python dictionaries by the Phenix GUI. The contents of the response dictionaries are generally log outputs that can be directly displayed in the GUI or a PDB model which can be read and interpreted as Phenix data structures. For example, when a sequence is sent to the webservice for an AlphaFold prediction, the sequence is sent to the Phenix-AlphaFold webservice as a text string, not as a sequence file. And when the GUI receives the predicted model, it is a text string in the JSON response, not as a file, and the model is processed into a Python object for subsequent calculations.
As an example of how the user would use the Phenix-AlphaFold webservice through the Phenix GUI, Figure 1a includes the input files from one of the tutorial datasets distributed with Phenix, the BAZ2a bromodomain based on PDB entry 7qz0 (Dalle Vedove et al., 2022). Figure 2 summarizes the flow of data between the user and the webservice. Once the user starts the job by clicking the “Run” button, the sequence, along with the parameters related to predicting the model (“Prediction strategy” and “Prediction Server” sections or “Prediction” button under “Advanced inputs” in Figure 1c), are sent to our server (step 1 in Figure 2) which then contacts the MMseqs2 server (Mirdita et al., 2019; Steinegger & Söding, 2017) (step 2 in Figure 2) to perform the multiple sequence alignment (MSA). This step is skipped if the exact sequence was submitted earlier. The cache is used to lessen the load on the MMseqs2 server, and the cache can be disabled by turning off the “Allow precalculated MSA” option in Figure 1c. The MSA cache is described in more detail in Section 4.
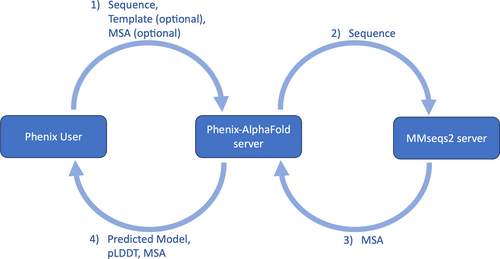
After the MSA is available and retrieved by our server (step 3 in Figure 2), we run AlphaFold to predict the model. The Phenix GUI retrieves the predicted model when it is available (step 4 in Figure 2), along with the predicted local distance difference test (pLDDT) scores and MSA. The user's computer will then iteratively rebuild and refine the predicted model against the experimental data. This refined model is then used as a template for the next cycle of structure prediction, rebuilding, and refinement. Figure 1d shows the overall log of the entire process. Figure 1e,f shows the statuses of the prediction and building/refining steps, respectively. The prediction status in Figure 1e provides information about whether or not the prediction was completed successfully, and the input information used for that prediction. For cycles after the first one, input information will include the MSA and the built model from the previous cycle as the template. Figure 1f summarizes the outcome of each cycle. Some important statistics for evaluating the cycle are the pLDDT scores that shows the confidence in the accuracy of the prediction, and the R-values (for diffraction data) that show how well the rebuilt/refined model fits that data. There is also a cache for results. If a user sends another job to the server with the same sequence and parameters, a cached result will be returned. The cache reduces the load on our server, and this cache can be disabled by turning off the “Allow precalculated results” option in Figure 1c. The results cache is described in more detail in Section 4.
Once the job completes, a final results panel, as shown in Figure 1g, will summarize the results, provide a listing of the final output files, and provide buttons for viewing the output in a molecular viewer like ChimeraX (Goddard et al., 2018) or Coot (Emsley et al., 2010). There is another button that can open a phenix.refine GUI in case further refinement is desired. For this particular tutorial dataset, the sequence is about 100 residues, and one prediction takes about 7 s to complete. The majority of the time for running the job is actually the rebuilding and refinement. That should take around 10 min on a contemporary computer.
More details about the implementation of the webservice, including software dependencies and hardware specifications, can be found in Section 4.
3 DISCUSSION
The Phenix-AlphaFold webservice is a new approach towards running Phenix programs and because of this, the webservice is limited to only running the AlphaFold prediction jobs that require more computational resources. As data about usage of the webservice grows, we will reevaluate this limit and potentially expand the kinds of jobs that can run on webservice to other Phenix programs. Moreover, if machine learning is successfully applied to other parts of the structure determination workflow, these can also be executed on the Phenix-AlphaFold webservice. This can be especially important if more large databases are required for future machine learning models or if some of the popular machine learning frameworks, like TensorFlow (Abadi et al., 2015) or PyTorch (Paszke et al., 2019), are more widely used in structural biology. TensorFlow is the underlying framework used for AlphaFold and PyTorch is used for OpenFold (Ahdritz et al., 2023), a structure prediction software package that is similar to AlphaFold. Both of these frameworks are optimized for NVIDIA GPUs and are usually run on Linux-based platforms. More integration of machine learning methods will most likely increase the computing requirements that initially spurred the development of the Phenix-AlphaFold webservice.
To that end, an important goal is providing a way to have reproducible servers with specific versions of the software. That is why, in the future, this webservice will be made available to the research community as an installer that has AlphaFold, and all of its dependencies included. The MMseqs2 server is also versioned and can be reproduced as well. This installer would allow users to host their own instance of the Phenix-AlphaFold webservice on their own hardware in a convenient way. The use of a conda environment will make it possible to track specific versions of all dependencies and deploy instances of the webservice with the same software stack. This will provide reproducible servers for specific versions of AlphaFold. Alternatively, this installer will also enable users to run the automated pipelines integrating AlphaFold on their Linux servers without having to set up a web server provided that the Linux server has the requisite GPU hardware and databases for AlphaFold. Being able to control the hardware also lets users who work with proprietary data to ensure that their data are kept secure. This is especially important to the commercial users of Phenix who prefer to not send any data outside of their networks. Lastly, this installer would be able to run on any Linux distribution without requiring Docker, the recommended way of installing and running AlphaFold according to the instructions from Google DeepMind (https://github.com/google-deepmind/alphafold). This simplifies system administration of the Linux server running the webservice because the installer avoids having to run an additional Linux distribution via Docker.
With this Phenix-AlphaFold webservice running on versioned conda environments, we are laying the groundwork that will enable Phenix developers to utilize the latest machine learning frameworks in future programs while providing a way for researchers to run these jobs remotely or locally in a consistent manner.
4 MATERIALS AND METHODS
The Phenix-AlphaFold webservice is running on a server with 64 cores (2 × AMD EPYC 7452), 1 TB of RAM, and 12 TB of SSD storage. There are three NVIDIA A100 GPUs, each with 40 GB of memory. The NVIDIA driver version is 470.57.02.
The software running the Phenix-AlphaFold webservice is CentOS 7.9 with Docker version 20.10.9 (build c2ea9bc). The base Docker image for AlphaFold 2.2 is running Ubuntu 18.04 with CUDA 11.1.1 (nvidia/cuda:11.1.1-cudnn8-runtime-ubuntu18.04 from DockerHub). The list of installed dependencies from Ubuntu are listed in the file “ubuntu_dependencies.txt” in the Supporting Information. We compiled httpd (httpd Developers, n.d.) version 2.4.58 for running the web server. For AlphaFold and Phenix, we use conda (Conda Contributors Conda, n.d.) to manage dependencies from conda-forge (Conda-Forge Community, 2015) and from pypi.org, the Python package index. The conda and pypi dependencies and versions are listed in the file “conda_environment.txt” in the Supporting Information.
We forked the official AlphaFold GitHub repository and modified the default Dockerfile so that the Docker image contains the additional dependencies for Phenix. The modifications are available at https://github.com/bkpoon/alphafold/tree/interactive and are added to version 2.2.2 of AlphaFold. After building the Docker image with the modified Dockerfile, we used the compiler in the Docker image to build a version of Phenix with code from April 28, 2023. By using the same conda environment for AlphaFold and Phenix, Phenix is able to access AlphaFold functions from Python.
We chose this approach because the instructions for installing and running AlphaFold calls for using Docker to build an image with the required dependencies (https://github.com/google-deepmind/alphafold?tab=readme-ov-file#installation-and-running-your-first-prediction). However, because the Docker image is based on Ubuntu 18.04, that can lead to security issues because Ubuntu 18.04 only has 5 years of free updates. To work around the use of Ubuntu 18.04 and to simplify system administration by removing Docker, we will switch to using a stand-alone Phenix-AlphaFold installer as described in Section 3. This change will help with the migration of our servers from CentOS 7 to Rocky Linux 9.
The REST server is configured as a Flask application (Flask Developers, n.d.) in httpd. The Flask application handles the conversion of the HTTP/JSON inputs into Python objects that can be used to perform the requested task. That is, when a user sends a POST request with a “start_job” Uniform Resource Locator (URL) and JSON payload to the server, Flask handles the parsing of both the URL and the JSON payload into the arguments needed to start the job with Python code in Phenix and AlphaFold. And when a user sends a GET request with a “get_job_result” URL and a JSON payload containing the unique job identifier, Flask will use that information to call the Python function to retrieve the job result associated with that job identifier and then format the output as a JSON response to the GUI.
The JSON payload for the “start_job” URL encapsulates the arguments provided to the program in the “args” key listed in Table 1. More specifically, the command-line arguments for the program that could be provided via the command-line are stored as a list of strings in the JSON payload. This provides the REST server the ability to start any Phenix command-line program in the future. In the current implementation, we are limiting the use to only phenix.predict_and_build and the list of all possible arguments can be shown on the command-line with “phenix.predict_and_build--show-defaults.” The Phenix GUI (Figure 1a–c) also shows what arguments can be changed. A full listing of all the parameters for Phenix 1.21 is also available on the phenix.predict_and_build documentation page (https://phenix-online.org/version_docs/1.21-5207/reference/predict_and_build.html). When the tool is updated, the latest documentation can be accessed from the Phenix website (https://phenix-online.org/documentation). The main arguments that are of interest to the user would be the sequence, and an optional model to serve as a template. For further customization, users can modify the other parameters presented in the GUI (e.g., the number of models to predict), but generally the defaults will work for most situations. An important note is that a token, included with Phenix 1.21 and later, is required for any interaction with the webservice. Phenix will automatically add the token to each JSON payload. Moreover, the Internet Protocol (IP) address of the user is also sent in the JSON payload for starting a job to help track usage.
When a job is submitted to the server, a directory is created with the unique job identifier as the name. There is a worker process that checks for newly created directories at regular intervals and starts the job if there are available GPU resources. If the server is full, jobs are queued and then run in the order that they are submitted as resources become available.
To help alleviate the load on the server, we cache the predicted models and the MSAs for 1 year. For predicted models, we use a hash based on the sequence and other parameters to determine uniqueness. For the MSA, we only use a hash of the sequence. When a new job is submitted, we calculate new hashes from the sequence and parameters. If the hash of the sequence and parameters match, we immediately return the predicted models from the cached job. If a parameter was changed, but the sequence hash is the same, we run the job, but use the cached MSA. The lifetime of the cache is 1 year, but a job that uses a cached result will reset the timer for that result. The user can turn off caching and have their result immediately deleted by turning off the “Allow precalculated results” option. The MSA cache can be disabled by turning off the “Allow precalculated MSA” option. These options are under the “Prediction Server” section in the GUI (Figure 1c) and their command-line equivalents are “allow_precalculated_result_file = False” and “allow_precalculated_msa_file = False” for disabling the results and MSA caches, respectively.
AUTHOR CONTRIBUTIONS
Billy K. Poon: Conceptualization; software; writing – original draft; writing – review and editing. Thomas C. Terwilliger: Conceptualization; software; writing – review and editing; funding acquisition. Paul D. Adams: Funding acquisition; writing – review and editing.
ACKNOWLEDGMENTS
We would like to acknowledge funding from the following sources. Lawrence Berkeley National Laboratory, grant DE-AC02-05CH11231 (PDA). National Institutes of Health grants, GM063210 (PDA, TCT) and GM141254 (BKP, PDA). The Phenix Industrial Consortium (BKP, PDA). We would also like to acknowledge Mirdata, Steinegger, Söding, and others who worked on MMseqs2 and providing the webservice for fast multiple sequence alignments.
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest to disclose.