Int&in: A machine learning-based web server for active split site identification in inteins
Review Editor: Nir Ben-Tal
Abstract
Inteins are proteins that excise themselves out of host proteins and ligate the flanking polypeptides in an auto-catalytic process called protein splicing. In nature, inteins are either contiguous or split. In the case of split inteins, the two fragments must first form a complex for the splicing to occur. Contiguous inteins have previously been artificially split in two fragments because split inteins allow for distinct applications than contiguous ones. Even naturally split inteins have been split at unnatural split sites to obtain fragments with reduced affinity for one another, which are useful to create conditional inteins or to study protein–protein interactions. So far, split sites in inteins have been heuristically identified. We developed Int&in, a web server freely available for academic research (https://intein.biologie.uni-freiburg.de) that runs a machine learning model using logistic regression to predict active and inactive split sites in inteins with high accuracy. The model was trained on a dataset of 126 split sites generated using the gp41-1, Npu DnaE and CL inteins and validated using 97 split sites extracted from the literature. Despite the limited data size, the model, which uses various protein structural features, as well as sequence conservation information, achieves an accuracy of 0.79 and 0.78 for the training and testing sets, respectively. We envision Int&in will facilitate the engineering of novel split inteins for applications in synthetic and cell biology.
1 INTRODUCTION
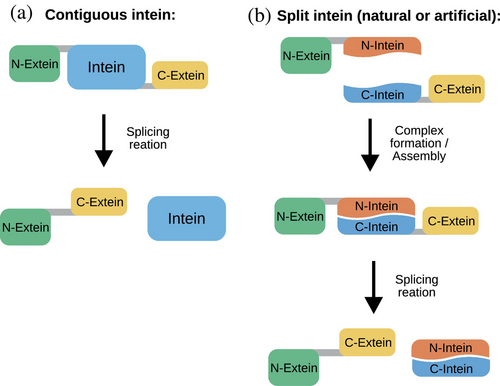
Inteins are small intervening proteins translated within host proteins that perform a so-called protein splicing reaction to excise themselves out of their flanking external polypeptides (exteins), which are ligated through a new peptide bond (Di Ventura & Mootz, 2019). Alongside contiguous inteins, which originate from a single gene (Figure 1a), split inteins (Figure 1b) encoded by two separate genes are of particular interest because they allow for a greater variety of applications (Waldhauer et al., 2015; Pan et al., 2016; Ye et al., 2018; Villiger et al., 2018; Lopez-Igual et al., 2019; Palei et al., 2019; Wang et al., 2019; Palanisamy et al., 2019; Purde et al., 2020; Choi et al., 2021). In the trans-splicing reaction, the N-terminal intein fragment (the ‘N-intein’) must first form a complex with the C-terminal intein fragment (the ‘C-intein’). After this step, the splicing reaction takes place resulting in the fusion of the N- and C-exteins (Figure 1b). In the past, inteins have been artificially split to obtain either a split intein out of a contiguous one (Mootz & Muir, 2002; Brenzel et al., 2006) or two intein fragments with low affinity for each other that could be induced to splice when in close physical proximity (Mootz & Muir, 2002; Tyszkiewicz & Muir, 2008; Yao et al., 2020). By controlling the physical proximity of the intein fragments with an external trigger such as light (Tyszkiewicz & Muir, 2008) or a chemical (Mootz & Muir, 2002), protein splicing is achieved in a conditional way, allowing interesting applications in synthetic and chemical biology (Mootz et al., 2003; Wong et al., 2015; Böcker et al., 2019) and cell biology (Lee & Muir, 2023). Alternatively, the intein fragments can be fused to proteins whose interaction one wishes to determine. If the proteins interact, splicing occurs leading to the accumulation of a splice product, such as a fluorescent protein, which can be easily quantified and is stable over time unless otherwise engineered (Yao et al., 2020). We call a split site allowing the splicing reaction to occur active, while one corresponding to two intein fragments unable to splice inactive.
So far, active split sites in inteins have been found with heuristic approaches (Wu et al., 1998; Mootz & Muir, 2002), or taking very simple protein structure considerations into account (Otomo et al., 1999). The existing SPELL algorithm (https://dokhlab.med.psu.edu/spell/) that was developed to computationally predict split sites in proteins for the construction of chemogenetic and optogenetic split proteins (Dagliyan et al., 2018), is inadequate for the specific case of inteins: we found that for ~80% of inteins collected from the literature (Aranko et al., 2014) no split sites could be predicted (33 inteins without any predicted split sites versus 8 with predicted sites (3D structures were predicted with AlphaFold2 (Jumper et al., 2021) as implemented in ColabFold (Mirdita et al., 2022); Source Data, File S1)). Moreover, for the split sites predicted to be active by SPELL, one site was experimentally shown to be active, while another to be inactive (Table S1). A different recently reported computational approach, ProteinSplit (http://elixir.fkkt.um.si/ProteinSplitIndex.html), is tailor-made for the prediction of ligand-mediated protein dimerization (Rihtar et al., 2023), and therefore unsuitable for the purposes of predicting active split sites in inteins. The ability to identify new active split sites in any intein of choice in an easy and confident manner would represent a breakthrough in the field and encourage a wider range of intein-based applications.
Here we apply machine learning (ML) to create an algorithm to predict active split sites in inteins. ML is a branch of artificial intelligence aiming to design algorithms that learn to recognize patterns in input datasets and make useful predictions on data not seen before. Given their power, ML algorithms started to pervade almost any research field, including molecular biology. ML approaches can be grouped into 4 different categories (Jovel & Greiner, 2021; Kouba et al., 2023): (1) supervised learning; (2) unsupervised learning; (3) semi-supervised learning; and (4) reinforcement learning. In supervised learning, models are trained on labeled data, where the connection between input and correct output is manually done to allow the algorithm to learn the connection and apply it later on a new, unlabeled dataset. Unsupervised learning models find hidden patterns or intrinsic structures in unlabeled input data. These models are particularly useful if labeled data are unavailable or too costly to obtain. In semi-supervised learning, the training data consist of a combination of labeled and unlabeled data. Reinforcement learning is a type of machine learning paradigm where the program learns to make sequential decisions to maximize cumulative rewards. The selection of the ML algorithm depends on diverse factors. If labeling data is a possibility, and the task is not about finding anomalies or reducing the dimensionality, then supervised learning is a natural choice. Lately, many ML algorithms have been put forward in the field of structural biology and protein design using a variety of learning methods (Khurana et al., 2018; Wang et al., 2019, 2023; Ferruz et al., 2022; Watson et al., 2023). Further information about the methodology, pitfalls and outlook of ML applications in protein engineering can be found in recent reviews (Villalobos-Alva et al., 2022; Kouba et al., 2023; Khakzad et al., 2023).
We used supervised learning to create an algorithm that combines several sequence and structural features, global or local in respect to the split site, and applies a logistic regression classification method on these features to predict the likelihood that a split site will be active. It moreover shows separation power with regard to the splicing efficiency, meaning it can be used to distinguish split sites that lead to fragments that reassemble efficiently and those that do not. The latter ones are particularly valuable for the creation of novel conditional split inteins. We validate the algorithm using data taken from the literature as well as generated by us and show that it has an accuracy of 0.79 for the training set and 0.78 for the testing dataset. The algorithm is freely available for academic research on the Int&in web server (https://intein.biologie.uni-freiburg.de/), while the source code of a standalone program is available on Github (https://github.com/bleblebles/Int-In/).
2 RESULTS
2.1 Creation of a dataset of active and inactive split sites in inteins
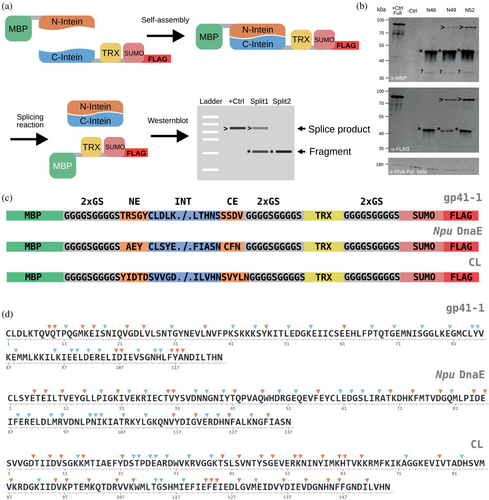
To train the machine learning model, we needed unbiased information not only on active sites—which are easily retrievable from the literature, but also on inactive ones. Therefore, we decided to generate a dataset of split sites using three inteins: gp41-1 (Carvajal-Vallejos et al., 2012), Npu DnaE (Iwai et al., 2006) and CL (a cysteine-less intein) (Bhagawati et al., 2019). To cover the whole sequence space of each intein, we randomly selected split sites to be experimentally tested with the only rule being that intein fragments had to be at least four residues long, because we reasoned that three or less residues would unlikely reassemble with the cognate fragment (Appleby et al., 2009; Mootz, 2009). The split sites were experimentally tested using Western blots as readout with antibodies allowing for the detection of the constructs made of the N-extein and the N-intein (N-construct), the C-intein and the C-extein (C-construct), as well as the splice product (Figure 2a,b, Source Data, File S2). As exteins we selected proteins known to be soluble in Escherichia coli: the maltose binding protein (MBP; N-extein), and thioredoxin (TRX) and SUMO with a FLAG tag (fused together; C-extein). To maximize the splicing reaction, we added the so-called “local exteins” (3–5 amino acids preceding the N-intein and 3–5 following the C-intein) (Lockless & Muir, 2009; Carvajal-Vallejos et al., 2012; Stevens et al., 2017; Bhagawati et al., 2019). These were separated from the exteins with a flexible linker (Figure 2c). We tested a total of 126 split sites (36 for gp41-1, 44 for Npu DnaE and 46 for CL) (Figure 2d). We considered active a site for which a band, albeit faint, could be detected at the size of the splice product. Of the 126 tested split sites, 64 were found to be inactive, and 62 to be active (Figure 2d). Efficiency of splicing varied across the split sites (Figure S1, and Source Data, File S3). Notably, inteins split at unnatural sites might exhibit a higher propensity for N- and/or C-cleavage, which are side-reactions of the main splicing reaction (Shah & Muir, 2013). Our design makes it difficult to assess this quantitatively, because the difference in size between the N-construct and the N-extein (i.e., the potential product of a N-cleavage) as well as between the C-construct and the C-extein (i.e., the potential product of a C-cleavage) is too small to be detected on a Western blot for many split sites. A different approach should be adopted to specifically account for N- and C-cleavage. Nonetheless, our calculations of splicing efficiencies at least indirectly reflect the presence of such side-reactions, which compete with the main splicing reaction and therefore inevitably lead to less splice product. We tried to indicate the presence of these side reactions as best as possible for split sites where the size difference allowed us to distinguish their products from the precursors (Figure S1).
2.2 Finding structural and biochemical properties with predictive power
Next, we sought to find evolutionary, structural and biochemical properties within inteins that may be helpful in discriminating between active and inactive split sites. These properties were extracted from the inteins' sequences and structures (for gp41-1 and Npu DnaE, crystal structures (PDB id: 6QAZ and PDB id: 4KL5, respectively), while for CL a structure generated through the ColabFold implementation (Mirdita et al., 2022) of AlphaFold2 (Jumper et al., 2021); Source Data, File S4). We considered the following properties: (i) binding affinity between the two resulting intein fragments, given its strong influence on whether a complex is formed or not (Figure 3a) (Vangone & Bonvin 2015, 2017); (ii) conservation of the residues around the split site (either based solely on sequence or based on spatial proximity in the 3D structure), as conserved regions are typically structurally or functionally important and should better not be tampered with (Figure 3a); (iii) relative surface accessibility of the residues around the split site, considering that residues exposed on the protein surface are likely to contribute less to overall protein stability than those residing in the core, and might thus be a good predictor for active split sites (Figure 3a); (iv) secondary structure elements, as regions with loops may contribute less to protein structure and could be favorable to locate split sites (Figure 3a); (v) affinity between the the first n residues of the C-intein and the full N-intein (what we call C-fragment docking) (Figure 3a). We considered this property having in mind the ‘capture and collapse’ mechanism introduced by Shah and colleagues, who investigated the naturally split Npu DnaE intein through NMR (Shah et al., 2013). The partially folded N-intein binds to the unfolded C-intein in a process termed capture, leading to an intermediate structure formed by the fully folded N-intein electrostatically bound by the still unfolded C-intein. During the collapse step, the C-intein folds, generating the functional split intein complex. Assuming generalizability of this mechanism, we decided to consider the C-fragment docking energy as a measure of how easily the intermediate structure of the intein forms.
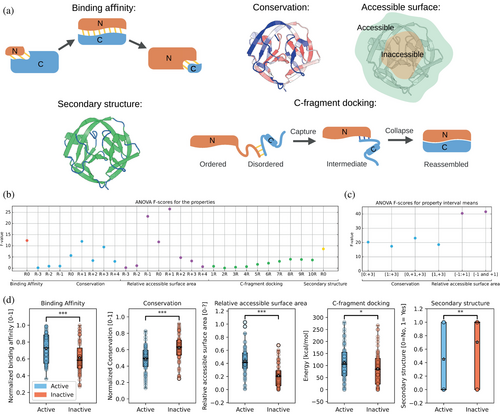
To evaluate the ability of each property to distinguish between active and inactive split sites, we calculated the F-value (Figure 3b), which is a metric that indicates how good the property is at correctly identifying active split sites, while minimizing mistakes. For the cases including additional residues beyond the split site itself, we calculated the F-value using the mean of the value of that specific property for different combinations of residues (Figure 3c). For ‘conservation’, we found that considering residues at positions 0, +1 and +3 gave the highest F-value (Figure 3c). Spatial conservation did not yield better results than sequence-based conservation (Figure S2; see Section 4 for the detailed explanation of how it was calculated). Therefore, the mean sequence-based conservation of residues at positions 0, +1 and +3 was finally used for this property. For ‘relative surface area’, residues −1 and +1 were used for the final property. For ‘C-fragment docking’, the highest F-value was obtained when including eight residues. All the tested properties were able to discriminate, in a statistically significant manner (Figure 3d), between active and inactive split sites, being, thus, promising for building the machine learning model.
2.3 Generating the machine learning model
Once we identified several features that can individually distinguish active from inactive split sites, we needed to build the decision-making algorithm—the classifier—which leverages these features to make predictions on unseen data. For selecting the most suitable one for our specific purpose of active split site identification in inteins, we compared several classifiers (Gaussian Naive Bayes, XGBoost, Logistic Regression, Decision Tree, and Support Vector Machine) with all possible feature combinations based on their performance measured by a 10-fold cross validation (10 CV) of the Matthews correlation coefficient (MCC) averaged over 10 runs (10 × 10-fold cross validation) (Figure 4a, Figure S3). The logistic regression classifier operating on all the features (namely, sequence conservation, relative accessible surface area, binding affinity, C-fragment docking energy and secondary structure) performed best (average 10 × 10 cross-validated MCC of 0.55, with standard deviation of 0.02). Therefore, it was used to generate the final model (Figure 4b, Table S2).
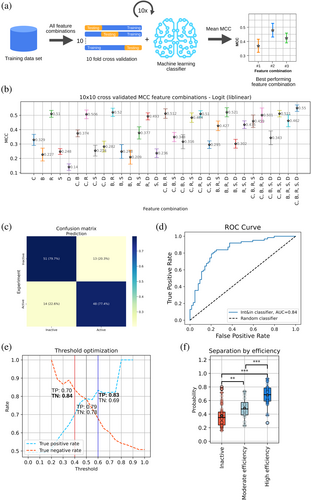
With the training dataset, the model had an MCC of 0.57, an accuracy of 0.79 (10-fold cross-validated accuracy of 0.784 ± 0.118), a precision of 0.79 and a recall of 0.77 (Figure 4c). The ROC (receiver operating characteristic) curve, which indicates the performance of a classification model at all classification thresholds in terms of true and false positive rates, had an area under the curve of 0.84 (Figure 4d). The model was able to correctly predict 82%, 78% and 76% of the active split sites for Npu DnaE, gp41-1 and CL, respectively (Figure S4).
The model relies on a threshold to define a split site as active or inactive. Considering that the probability p is a value from 0 to 1, we set the default classification threshold at 0.5, so that any split site with a predicted probability p ≥ 0.5 is classified as active, while those with p < 0.5 are classified as inactive. With this threshold, the true positive and negative rates are 0.79 and 0.78, respectively. The threshold can be, however, adjusted to specifically increase one of these rates, something that might be required in specific cases (Figure 4e). By employing two different classification thresholds, the true positive or true negative rate can be individually increased. As changing the classification threshold also reduces the number of predicted split sites, we set the limit for these thresholds so that the number of predicted split sites does not go below half of the total number of experimentally validated active/inactive sites. Setting the threshold to 0.6, the true positive rate can be increased from 0.79 to 0.83; setting it to 0.4, the true negative rate can be increased from 0.78 to 0.84. Using these two thresholds, we created four categories of prediction certainty: active split sites (p ≥ 0.5), inactive split sites (p < 0.5), active with high probability (p ≥ 0.6), and inactive with high probability (p < 0.4).
Next, we were interested in knowing whether the model could be used to differentiate split sites characterized by lower or higher splicing efficiency (Source Data, File S2). To this end, the probability of a site to be active, as outputted by the model, was plotted for three groups: inactive split sites, split sites with moderate splicing efficiency (<50% of the splicing efficiency) and split sites with high splicing efficiency (≥50% of the splicing efficiency). We found a significant difference between inactive and moderate efficiency sites, as well as between moderate and high efficiency sites (Figure 4f).
2.4 Model validation
To assess the performance of the model, we extracted from the literature a dataset of 97 split sites in inteins, of which 57 active and 40 inactive, from 41 different inteins (Aranko et al., 2014) (Table S3). We excluded Pfu RIR1-1 and Pfu RIR1-2, as they were split inside their homing endonuclease/stirrup domain, as well as gp41-1 and Npu DnaE, since they are already contained in the training dataset. Given the presence of different inteins in this dataset, we reasoned it was well suited to evaluate the generalizability of the model, which was obtained with data from three inteins. It is important to note that this testing dataset, even though quite diverse from the point of view of the variety of inteins included, is unbalanced due to the prevalence of split sites clustering around the naturally occurring ones, as well as to the over-representation of active split sites (so-called class imbalance; 40 inactive vs. 57 active sites).
We generated the 3D structures of all the inteins with AlphaFold2 (Jumper et al., 2021) (implementation in ColabFold (Mirdita et al., 2022); Source Data, File S1), inputted them into the model, and then calculated performance measures (Figure 5a,b). We found an MCC of 0.55, an accuracy of 0.78 (10-fold cross-validated accuracy of 0.732 ± 0.152), a precision of 0.76, a recall of 0.91 and a ROC of 0.83. Applying the same thresholds used with the training dataset, the true positive rate could be increased to 0.85 from 0.76, while the true negative rate decreased slightly from 0.83 to 0.82 (Figure 5c). The latter may be due to the imbalance of the dataset mentioned above.

To check for the effect of class imbalance, we employed an undersampling strategy based on the NearMiss (version 3) technique (Lemaître et al., 2017), which selects samples from the majority class based on their closeness to samples in the minority class. Using this undersampled testing dataset, we obtained an MCC of 0.62, an accuracy of 0.79 (10-fold cross-validated accuracy of 0.75 ± 0.158), a precision of 0.71, and a recall of 0.98 (Figure S5). Together, these results show that the logistic regression-based model performs similarly with the literature-derived validation dataset as with the training dataset containing sites spanning the whole sequence space of three inteins.
2.5 The Int&in web server
To allow researchers of varying levels of computational knowledge to make use of this model, we opted for making it available through a freely-accessible web server (https://intein.biologie.uni-freiburg.de), which is free of charge for academic usage. The Int&in web server evaluates each amino acid of a given intein sequence for its potential to be an active split site, and provides an easy-to-use web interface to quickly evaluate the results. Users can anonymously upload their experimentally determined or modeled protein structures and receive a personalized link to visualize the results. Additionally, we provide the option to submit batch runs with multiple .pdb files. Finally, users can register for an account, which ensures that past jobs are easily retrieved and can be analyzed at a later point (past jobs may be deleted after 30 days). After all calculations are executed on the backend, usery are notified via email (if an address was provided at job submission; otherwise, the job can be accessed through the personalized link that appears after job submission), and can access an interface consisting of a structure window, a list of split sites and several graphs showing the values of the individual properties as well as the model predictions. All files generated by the Int&in web server as well as the raw data from third party programs (conservation calculations and secondary structure calculations) can be downloaded.
3 DISCUSSION
Here we described Int&in, a web server that relies on a machine learning algorithm to predict active and inactive split sites in inteins. Before embarking in the development of this web server and the model it relies on, we checked the literature and found SPELL, a web server for the prediction of split sites in proteins specialized in the task of protein functional reconstitution by means of ligand- or light-regulated heterodimerizing systems (Dagliyan et al., 2018). Indeed, locating functional split sites in proteins is a highly desirable task, because splitting proteins into two dysfunctional halves, which can be brought back into close physical proximity to regain activity, is a useful technique in cell and synthetic biology to control and/or understand biological processes (Cali & Brini, 2021; Mahameed et al., 2022; VarnBuhler et al., 2022). The algorithm behind SPELL makes use of an energy function and several structural and sequence-based parameters to determine functional split sites. Because it was generated from a limited data set of 27 functional split sites from 16 different proteins, it is difficult to assess whether the rules employed by SPELL truly reflect the full range of sites at which a protein can be split. This was not the main goal of the SPELL algorithm, which aims to maximize the number of true positive sites while keeping the number of false positive sites at a minimum. This means that several functional split sites that are not deemed optimal by the algorithm are lost, which can be problematic when wishing to split a protein in a specific region. For inteins in particular, the possibility to find split sites in specific regions can be crucial to, for instance, generate very short intein fragments, which can be more easily chemically synthesized and used in protein semisynthesis (Ludwig et al., 2006; Mootz, 2009; Burton et al., 2020) or be more amenable to control via caging within the light-sensitive light oxygen voltage (LOV) domain (Wong et al., 2015). Interestingly, when applied to 41 individual inteins, SPELL predicts no split sites for around 80% of the inteins tested (Table S1). Additionally, when we applied the same energy profile used by SPELL, which is conceptually equivalent to the binding energy we use, we found no significant difference between inactive and active split sites (Figure S6a,b). This indicates that SPELL is not particularly suitable to predict active split sites in inteins. Very recently, another algorithm, ProteinSplit, has been developed to specifically predict how to split a protein of interest for functional reconstitution via a ligand (Rihtar et al., 2023). Since this model requires a ligand, it is not immediately applicable to the vast majority of inteins, which spontaneously splice and are not ligand-dependent.
Our model uses binding affinity between the resulting intein fragments, as well as conservation, relative accessible surface area, C-fragment docking energy and secondary structures of and around the split site to calculate a probability score. Trained on randomly distributed split sites from three different inteins amounting to 126 sites in total, the model achieved an accuracy of 0.78 with an untrained dataset consisting of 41 different inteins with split sites gathered from literature. Given their reliance on the training dataset to learn the patterns in the data, it is not surprising that machine learning algorithms strongly depend on the size of the training dataset, with larger ones leading to better predictive power (Sordo & Zeng, 2005). We were pleased to see that our model performs well despite the modest size of the training dataset.
As the model was not trained with any knowledge of the efficiency of splicing of each site, but nonetheless managed to output probabilities that show a significant difference between split sites associated with moderate and high splicing efficiencies, we speculate that the properties used in the model are also predictive of splicing efficiency associated with each split site, and thus Int&in could be used also to predict efficiencies. By testing the model on a dataset coming from the literature, characterized by a variety of inteins with different exteins expressed in different model organisms, we have shown generalizability as well as wide applicability of our model. Nonetheless, in cases where the local exteins are changed or truncated the model's output may show discrepancies, considering the model was trained on results generated with the optimal local exteins for each intein.
Additionally, we would like to encourage researchers to share the “negative data” (in the case of inteins, split sites that result in non-splicing inteins) as well, since such data would be equally informative as “positive data” and would prevent class imbalance issues in future ML algorithms' development. While the presence of class imbalance was not problematic for our particular application, it might be quite deleterious in others. In general, training ML models on diverse and large datasets would result in improved generalization, reduced overfitting, enhanced model complexity, improved accuracy and stability, better data representation and better learning performance.
The source code for a standalone version of the tool as well as the files for feature selection, model training and testing can be accessed at https://github.com/bleblebles/Int-In/. Since the Int&in web server is user-friendly, and requires no prior knowledge of bioinformatics or computational biology, we believe it will boost research on these fascinating proteins as well as applications based on them.
4 MATERIALS AND METHODS
4.1 Non-covalent bond determination in Int&in
A list of residues that are in proximity to each other (10 Å at most) is created and the existence of non-covalent bonds (hydrogen bond, salt bridge, van der Waals, pi-pi and pi-cation) is determined, as outlined below.
4.2 Hydrogen bonds
A hydrogen bond between two atoms is considered to be present if the angle between the hydrogen donor (D) and the hydrogen acceptor (A) is ≤63° and the distance between them is ≤3.5 Å (40). N, O, S and atoms may be acceptors and/or donors with the exception of NH3, which cannot be an electron acceptor. Moreover, C atoms may be donors, to account for backbone hydrogen bonds (Gu et al., 1999). Donor atoms additionally need at least one hydrogen atom covalently bound to them.
4.3 Salt bridges
A salt bridge between two residues is considered to be present if they have opposite charge and the distance between any of the nitrogen atoms in the side chain of the positively charged residue and the oxygen atoms (or the sulfur of cysteine) in the side chain of the negatively charged residue is ≤4 Å (Barlow & Thornton, 1983).
4.4 Aromatic interactions
π–π interactions are attributed to aromatic amino acids if the centroids of their aromatic rings are at most 7.5 Å from each other and none or only one of the aromatic rings is positively charged (Bhattacharyya et al., 2002). Cation-π interactions are considered to be present if the distance between the centroid of the aromatic ring of a neutral aromatic amino acid and any nitrogen atom of the side chain of a positively charged lysine or argine is ≤6 Å (Gallivan & Dougherty, 1999).
4.5 van der Waals
A van der Waals interaction is considered to be present between any carbon–carbon, carbon–sulfur, carbon–oxygen (glutamine OE1 and asparagine OD1) and carbon-nitrogen (glutamine NE2 and asparagine OE1) if their distance is ≤0.5 Å (Lovell et al., 1999). The van der Waals radii (NACCESS radii) of the two potentially interacting atoms is subtracted from the distance between the atoms to get the distance between the atoms' surfaces (Hubbard et al., 1993).
4.6 C-fragment docking energy determination in Int&in
The docking energy between the first eight residues of the C-intein and all residues of the N-intein is calculated by attributing different energies to each non-covalent interaction depending on its type (and distance, in the case of hydrogen bonds):
| Non-covalent interaction | Energy (kJ/mol) |
|---|---|
| Hydrogen bond | For distances between D and A ≤ 1.5 Å: 115.0 For distances between 1.5 Å < D and A ≤ 2.2 Å: 40.0 For distances between 2.2 Å < D and A ≤ 3.5 Å: 17.0 |
| Salt bridge | 20.0 |
| Aromatic interaction | Pi–pi interaction: 9.4 Pi–cation interaction: 9.6 |
| van der Waals | 6.0 |
The energies have been used according to the RING 3.0 web server (Clementel et al., 2022).
4.7 Relative accessible surface area determination in Int&in
4.8 Conservation determination in Int&in
4.9 Spatial conservation determination
Spatial conservation was calculated based on mean, minimum and maximum conservation values of all residues that have a specified maximum distance from the split site. For this, Cα-atom positions were used, while the split site position was denoted as the middle of Cα-atom positions of the two flanking residues at the split site.
4.10 Binding affinity determination in Int&in
The binding affinity between two fragments is calculated through the PRODIGY prediction model established by Vangone and Bonvin (2015, 2017). The model is implemented in the backend of the Int&in web server by refactoring the code from Python to C#. The surface accessibility is calculated through an implementation of the algorithm by Lee and Richards (1971) and Mitternacht (2016) as outlined in the FreeSASA source code, with 20 sphere slices and the van der Waals radii and maximally accessible surface areas as defined by the NACCESS program (Hubbard et al., 1993).
4.11 Secondary structure determination in Int&in
The secondary structure of a sequence is calculated with the DSSP program (Kabsch & Sander, 1983; Joosten et al., 2011). A split site is considered to be in a secondary structure if both flanking residues are part of a secondary structure.
4.12 Computational resources needed for Int&in
- PDB2PQR v2.1.1—to generate the structure at a pH of 7 and to add hydrogens
- DSSP (3.0.0)—to generate the secondary structure
- HMMER (3.3)—to identify homologous proteins in the UniRef90 database (2021_03)
- Muscle (v3.8.1551)—to create a multiple sequence alignment (MSA) of the sequence contained in the .pdb file submitted by the user and maximum 2000 sequences extracted from the HMMER output
- Rate4Site (3.0.0.)—to generate the conservation score out of maximum 150 clustered sequences from the MSA
- Python, with the following two libraries: scikit-learn (1.1.0) and pandas.
The web-based GUI makes use of the following JavaScript libraries: jQuery (https://github.com/jquery/jquery), NGL viewer for molecular representation (Rose & Hildebrand, 2015) and dygraphs for chart plots (https://github.com/danvk/dygraphs). The web-based GUI also lets the user create an account to have an overview of the submitted jobs. The account information is stored in a MYSQL database. Passwords are hashed for security.
4.13 Property significance tests
To assess if the difference between the group of split sites predicted to be active and that of split sites predicted to be inactive was significant when using a defined property, we applied either the t-test or the Mann–Whitney U test (scipy.stats.ttest_ind, mannwhitneyu), depending on whether the groups were considered normally distributed or not, respectively. Normal distribution was evaluated using the Shapiro–Wilk test, the normaltest function from scipy and the Anderson-Darling test (scipy.stats.shapiro, normaltest, anderson).
4.14 Property optimization with ANOVA F-scores
The ANOVA F-scores were calculated with the sklearn library in Python.
(sklearn.feature_selection.SelectKBest, f_classif ).
4.15 Feature combination selection
Different combinations of features were evaluated on different models (from sklearn.naive_bayes.GaussianNB, xgboost.XGBClassifier, sklearn.linear_model.LogisticRegression, sklearn.tree.DecisionTreeClassifier, and sklearn.svm.SVC) by calculating the 10x Cross-validated (sklearn.model_selection.KFold with shuffle set to true) Matthews correlation coefficient (MCC, sklearn.metrics.matthews_corrcoef), which was then averaged over 10 runs with different seeds (from numpy.random.seed, seeds 0–9 were used).
4.16 Model creation
The logistic regression model (from sklearn.linear_model.LogisticRegression (Pedregosa et al., 2011); with the ‘liblinear’ solver, which was selected due to the smaller dataset), was trained in Python on the training dataset. To evaluate the model in terms of accuracy, precision, recall, and MCC confusion matrices were generated (sklearn.metrics.accuracy_score, precision_score, recall_score, matthews_corrcoef, confusion_matrix). The confusion matrix representation was generated through seaborn (Waskom, 2021). The model was saved through the pickle library in Python.
4.17 Protein structures
The crystal structures of gp41-1 and Npu DnaE (PDB id: 6QAZ and PDB id: 4KL5, respectively) were used. The structures were additionally modified in PyMol to remove any extein sequences and mutate residues so that they would represent the native structures. For the gp41-1 intein structure, the first three extein residues (SGG) were removed and the fourth alanine (which inactivates the intein) was mutated back to cysteine. For Npu DnaE chain A was used for the computational experiments and the first three extein residues were also removed and the fourth alanine residue was mutated to cysteine; additionally, the last four extein residues (ADNG) in the structure file were also removed. CL as well as all other inteins from the literature data set were modeled through the ColabFold implementation (Mirdita et al., 2022) of AlphaFold2 (Jumper et al., 2021).
4.18 Plasmid construction
For the expression of all intein constructs in E. coli, the pTrc99A vector was used. A list of all used primers and amino acid sequences of exemplary plasmids is given in the Appendix, File S5. For PCR amplification the Phusion Flash High-Fidelity PCR Master Mix (2×) from ThermoScientific and the Biometra TOne 96G thermocycler from Analytik Jena were used. Plasmids were constructed using the NEBuilder® HiFi DNA Assembly Cloning Kit from New England Biolabs.
The mbp gene coding for the maltose binding protein (MBP) was amplified from pETM41 via PCR with primers pTrc_MBP_fw and MBP_gp41_rev (Figure S7a, upper left panel). Note that the gene in the final plasmids contains a mutation in the first base of the second codon, which means that the protein has lysine instead of glutamic acid at that position. Since the anti-MBP antibody worked well we decided against removing the mutation. The trx gene coding for thioredoxin (TRX) was amplified from pETTrx with primers TRX_fw and TRX_SUMO_rev (Figure S7a, upper middle panel). Plasmids pETM41 and pETTrx were a kind gift of Gunter Stier (Heidelberg University). The Saccharomyces cerevisiae smt3 gene coding for SUMO was amplified from pTB324_AmiB (kind gift of Thomas Bernhardt, Harvard Medical School, Boston) with primers SUMO_fw and SUMO_rev (Figure S7a, upper right panel). The N- and C-fragments of the gp41-1 gene were amplified with primers gp41-1_fw and gp41-1_N_merge_rev, and primers gp41-1_rev and gp41-1_C_merge_fw, respectively, from pSiMPlk (Palanisamy et al., 2019) (Figure S7a, lower left panel). The backbone fragments were amplified with primers pTrc_BB_rev and CoLE1_fw as well as CoLE1_rev and SUMO_BB_fw from pTrc99A in order to have shorter fragments. All fragments (backbone 1, backbone 2, mbp, gp41-1 N-fragment, gp41-1 C-fragment, trx, sumo; Figure S7b, upper panel) contain overhangs allowing for assembly yielding pTRc-MBP-gp41-1-TRX-GS-SUMO-FLAG (Figure S7b, lower left panel). Subsequently, GS linkers (one GS linker consists of the following sequence: GGGGSGGGGS) were added upstream of the local exteins of the N-intein and downstream of the local exteins of the C-intein (Figure 2c) in pTRc-MBP-gp41-1-TRX-GS-SUMO-FLAG plasmid with primers MBP_GS_rev and CoLE1_fw, Intein_GS_TRX_fw and CoLE1_rev as well as GP41_GS_Nextein_fw and GP41_Cextein_rev yielding pTRc-MBP-GS-gp41-1-GS-TRX-GS-SUMO-FLAG (Figure S7b, lower right panel). pTRc-MBP-GS-GS-TRX-GS-SUMO-FLAG containing the N- and C-exteins and the native local extein sequences for gp41-1 (positive control) was generated from pTRc-MBP-GS-gp41-1-GS-TRX-GS-SUMO-FLAG using primers GS_GP_N_and_C_Ext_fw, CoLE1_rev and GS_Next_rev and CoLE1_fw.
The constructs with Npu DnaE and CL were constructed using pTRc-MBP-gp41-1-TRX-GS-SUMO-FLAG as template. The gene coding for full-length Npu DnaE was amplified with primers MBP_GS_rev and CoLE1_fw, Intein_GS_SUMO_fw and CoLE1_rev as well as Npu_CExtein_rev and Npu_NExtein_fw yielding pTRc-MBP-GS-Npu_DnaE-GS-TRX-GS-SUMO-FLAG. The gene coding for CL was amplified from pTT32 and pTT43 (kind gift of Henning Mootz, University of Münster; Bhagawati et al., 2019). Specifically, the N-intein was amplified from pTT32 with primers AesN_GS_fw and AesN_rev, while the C-intein was amplified from pTT43 with primers AesC_fw and AesC_GS_rev. The two backbone fragments were amplified from pTRc-MBP-GS-gp41-1-GS-TRX-GS-SUMO-FLAG with primers MBP_GS_rev and CoLE1_fw, Intein_GS_SUMO_fw and CoLE1_rev. The four fragments were then assembled into pTRc-MBP-GS-CSIntein-GS-TRX-GS-SUMO-FLAG.
To clone the different split sites, we generated a so-called ‘split cassette’, containing a stop codon, a frame-shifted stop codon followed by a random spacer DNA, a ribosome binding site (Elowitz & Leibler, 2000) and a start codon. This split cassette was inserted in the intein-containing plasmid via two overlapping primers encoding the split cassette in their overhangs and annealing to the sequence of the intein at the respective split site (see Source Data, File S5 for a list of representative and unique plasmid sequences and a full list of primers. The split cassette primers contain the name of the respective intein and the split site). Two primers annealing to the backbone (CoLE1_fw and CoLE1_rev) were used to generate two fragments with the primers containing the split cassette. Note that all constructs contain the same exteins and the same flexible linkers (GGGGSGGGGS) that separate them from the local exteins. The only difference among constructs is the local exteins, which are specific to the intein (Figure 2c).
4.19 Bacterial cell lysis
Individual colonies were picked and used to start overnight (ON) cultures, which were grown at 37°C with 250 rpm shaking in the multitron pro incubator (Infors AG). The next morning, a volume of (100/OD) × 3 μL of the ON culture was used to inoculate a fresh tube with 3 mL LB medium plus ampicillin (100 mg/L). The tubes were subsequently shaken at 37°C and 250 rpm for 90 min, after which 3 μL of 1 M Isopropyl β-d-1-thiogalactopyranoside (IPTG) were added to each tube. The tubes were then put again inside the incubator and shook for 2 h and 30 min at 37°C 250 rpm. Afterwards, the OD600 of each culture was measured and a volume of 100/OD μL was taken and centrifuged at 13,000 rpm for 4 min at room temperature. The supernatant was removed from the samples, and the pellets resuspended in 20 μL 4× Lämli Buffer (Bio-Rad) and 80 μL ddH2O. The tubes were heated up for 10 min at 95°C and stored at −20°C. The OD at 600 nm of the bacterial cultures was measured with the OD600 DiluPhotometer from IMPLEN GmbH.
4.20 Western blot
1.5 μL of each cell extract were loaded in a well of a Mini-PROTEAN® 10% TGX™ Precast Gel (10 wells, 50 μL pocket volume; Bio-Rad) and separated in the Mini-PROTEAN Tetra Vertical Electrophoresis Cell (Bio-Rad) at 100 V for around 1 h and 30 min. Protein transfer onto a PVDF membrane was carried out with the Trans-Blot Turbo Mini 0.2 μm PVDF Transfer Packs (Bio-Rad) and Trans-Blot Turbo Transfer System (Bio-Rad). The membrane was blocked for 2 h with 5% BSA-PBST solution at room temperature on a rocking machine at 25 rpm. The blocking buffer was removed and 10 mL 5% BSA-PBST with 1 μL RABBIT anti-DYKDDDDK Tag antibody (Bio-Rad, # AHP1074), 1 μL anti-MBP monoclonal antibody (New England Biolabs, #E8032L) and 6 μL anti-E. coli RNA Polymerase β-subunit antibody (BioLegend, #663905) were added. The membrane was then placed on a rocking machine at 25 rpm for another 2 h. The BSA-PBST antibody mixture was then removed and the membrane was washed three times with PBST with gentle rocking for 5 min. 4 μL of the Cy5 goat anti-rabbit antibody (Invitrogen, #A10523) and 4 μL of the Alexa Fluor goat anti-mouse antibody (Invitrogen, #A11029) were added to 10 mL BSA-PBST and poured over the membrane in a closed box (used to protect it from light). The box containing the membrane was placed on the rocking machine at 25 rpm for another hour. Subsequently, the secondary antibody mixture was removed followed by three washes with PBST, during which the membrane was rocked for 5 min in the dark box. The membranes were imaged with the Amersham Typhoon 5 (Global Life Sciences Solutions) with the Cy2 and Cy5 emission filters and excitation wavelengths.
4.21 Calculation of splicing efficiencies
4.22 Definition of experimentally validated active split site
Two independent WBs were performed for each split site for each tested intein. A split site was considered active only if the splice product was quantifiable in both replicates.
AUTHOR CONTRIBUTIONS
Mirko Schmitz: Methodology; software; writing – original draft; formal analysis; data curation; investigation; validation; visualization. Jara Ballestin Ballestin: Validation; methodology; data curation. Junsheng Liang: Methodology; validation; data curation. Franziska Tomas: Methodology; validation; data curation. Leon Freist: Software. Karsten Voigt: Software. Barbara Di Ventura: Writing – review and editing; supervision; funding acquisition; resources; formal analysis. Mehmet Ali Öztürk: Conceptualization; supervision; formal analysis.
ACKNOWLEDGMENTS
This work was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement no. 101002044 to BDV), and partly by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project #422681845 within SFB1425. Open Access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST STATEMENT
The authors declare that there are no competing interests.