PBITV3: A robust and comprehensive tool for screening pathogenic proteomes for drug targets and prioritizing vaccine candidates
Shuvechha Chakraborty and Mehdi Askari to be considered as joint first authors.
Review Editor: Nir Ben-Tal
Abstract
Rise of life-threatening superbugs, pandemics and epidemics warrants the need for cost-effective and novel pharmacological interventions. Availability of publicly available proteomes of pathogens supports development of high-throughput discovery platforms to prioritize potential drug-targets and develop testable hypothesis for pharmacological screening. The pipeline builder for identification of target (PBIT) was developed in 2016 and updated in 2021, with the purpose of accelerating the search for drug-targets by integration of methods like comparative and subtractive genomics, essentiality/virulence and druggability analysis. Since then, it has been used for identification of drugs and vaccine targets, safety profiling of multiepitope vaccines and mRNA vaccine construction against a broad-spectrum of pathogens. This tool has now been updated with functionalities related to systems biology and immuno-informatics and validated by analyzing 48 putative antigens of Mycobacterium tuberculosis documented in literature. PBITv3 available as both online and offline tools will enhance drug discovery against emerging drug-resistant infectious agents. PBITv3 can be freely accessed at http://pbit.bicnirrh.res.in/.
1 INTRODUCTION
The emergence of multi-drug resistant pathogens necessitates the need to increase the repertoire of anti-infective agents and their targets. Although, several approaches such as comparative and subtractive genomics, essentiality/virulence and druggability analysis are used for novel target prediction; a software that integrates the workflow for high throughput screening and analysis was lacking.
This prompted our team to develop an online webserver named Pipeline Builder for Identification of Targets (PBIT), in 2016, which incorporated several in silico approaches to screen microbial proteomes for high-throughput prediction of drug targets such as non-homology analysis against human proteome, anti-target, and gut-microbiota; essentiality and virulence analysis, druggability analysis and determination of functional and pathway attributes of these targets (Shende et al., 2017). Recently, topological network analysis and metabolic flux analysis using genome-scale metabolic models (GSMMs) have gained importance in prioritizing targets in pathogens like Bacillus cereus, Mycobacterium tuberculosis, Plasmodium falciparum, Klebsiella pneumonia, etc. (Anis Ahamed et al., 2021; Zhu et al., 2022) Therefore, we developed an offline version of PBIT (v2) in 2021, wherein the pipeline was extended to incorporate network-based and metabolic systems biology-based approaches in target identification. The application of offline PBIT modules was confirmed by validating targets identified from Candida albicans and Candida tropicalis proteomes using published literature and in vitro methods (Mukherjee et al., 2021).
For pathogens that significantly affect human health, development of vaccines by reverse vaccinology is an attractive option. Given the widespread availability of genomic data for many pathogenic organisms, sequence-based screening for antigenic features is a feasible approach. Hence, we have now developed PBITv3; wherein, an immuno-informatics module has been introduced that can screen target sequences based on its antigenicity or ability to mount B-cell or T-cell based immune response. We have also updated the background databases and algorithms in PBITv3 for additional functionalities (Table 1).
| Modules | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comparative genomics | Annotation | Systems biology | Immunoinformatics | ||||||||||||
| Sub-modules | Non-homology against | Essentiality and virulence analysis | Druggability | Host–pathogen interaction | Broad-spectrum analysis | Function and subcellular localization | KEGG pathway (pathogen vs human) | Topological network analysis | Reaction essentiality analysis (FVA) | In silico gene knockout | Prediction of | ||||
| Human proteome | Human anti-target | Gut-microbiome | T-cell epitope | B-cell epitope | Antigenicity | ||||||||||
| Data source/sequence count (n)/algorithms | |||||||||||||||
| Application type | UniProtKB proteome | Review of articles | Review of articles | DEG, VFDB, DFVF | DrugBank, TTD | HPIDB, PHISTO, PHIbase | Review of articles & UniProt | UniProt | KEGG | iGraph package (R 4.0.3) | Cobrapy (Python 3.6) | NetMHCpan 4.1 | Multiple algorithms | Protengen db, Vaxijen | |
| Online (Shende et al., 2016) | n = 70,959 | n = 296 | n = 4732 from 83 species | n = 78,029 | n = 848 | n = 4371 | Proteome of 180 pathogens | - | - | Absent | |||||
| Offline (Mukherjee et al., 2021) | n = 70,244 | - | - | - | n = 8372 | - | - | - | - | - | - | - | Absent | ||
| Online and offline (this paper, 2023) | n = 42,454 | n = 484 | n = 504,580 from 147 species | n = 75,458 | n = 18,002 | n = 17,775 | Proteome of 520 pathogen | - | - | - | - | - | - | - | - |
| Remarks for current update | Removed TrEMBL sequences | Removed non-validated sequences | Included ChEMBL data for small molecules | MHC I and MHC II prediction | Refer main text | Alignment based and alignment free prediction | |||||||||
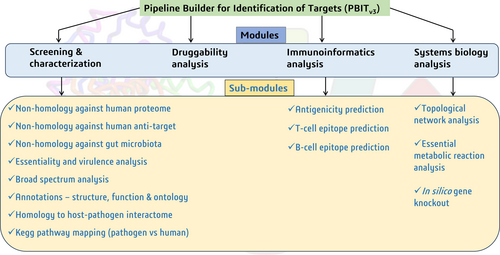
2 FRAMEWORK AND FUNCTIONALITIES
PBITv3 is available as web-based as well as command line-based tool that is compatible with Windows 10 and above. The tool was developed using PERL(v5.32), BioPerl (v1.007001), Python (v3.7) and R (v4.0.3) and BLAST+ 2.13.0 executables. A brief description of all the modules in PBITv3 (Figure 1) is given below.
2.1 Screening and characterization module
Using this module, proteome of pathogens (up to 500 sequences) can be concurrently screened using subtractive genomics methods, and subsequently annotated based on sequence similarity with curated databases. The sequence similarity of the input sequences to queried databases is computed using BLASTp.
2.1.1 Non-homology against human proteome, human anti-target, gut microbiota
These sub-modules help to screen out sequences that share close sequence similarities with human proteome (UP000005640), anti-targets and gut microbiome. The human proteome consists of 42,432 protein sequences as per UniProt Proteomes (TU Consortium, 2023) (as on August 28, 2023); of which 20,408 are canonical sequences and rest are isoforms. Anti-targets are human proteins that can trigger unwanted side effects under the influence of a drug and hence should not be targeted. This list has been compiled from literature (Kowalska et al., 2020; Lagunin et al., 2018; Zianna et al., 2022; Garcia-Sosa, 2018; Cavalluzzi et al., 2020) and consists of 484 protein sequences. The database for gut microbiota consists of referenced proteome sequences (504,580 sequences) from UniProt and RefSeq databases of 147 microbes curated from literature (Appendix S1).
2.1.2 Essentiality and virulence analysis
An essential or virulent protein are crucial for pathogen's survival or pathogenicity. Through PBIT, such proteins can be predicted based on sequence similarity to essential proteins in other bacteria, eukaryotes or archaea sourced from Database of essential genes (DEG v15) (Luo et al., 2021), and virulent proteins (sources: DFVF (Database of fungal virulence factors)) (Lu et al., 2012) and VFDB (Virulence factor database) (Liu et al., 2022).
2.1.3 Broad spectrum analysis
This sub-module helps to identify poly-microbial drug targets that have homologs in multiple pathogens. These targets are important for development of broad-spectrum drugs to treat multiple infections. The database for broad-spectrum analysis comprises of UniProt referenced proteomes of 520 pathogens (except commensals) as categorized by CDC (Centre for disease control) on January 1, 2023.
2.1.4 Homology to host–pathogen interactome
Pathogen proteins that interact with host play an important role in infection, invasion and induction of host immune response. Such proteins are ideal targets for therapeutic interventions. This sub-module helps to shortlist proteins that share sequence similarity with microbial proteins that are involved in host interaction, based on the data available in Host–Pathogen Interaction Database (HPIDB) 3.0 (Ammari et al., 2016), Pathogen–Host Interaction (PHI)-BASE 4.15 (Urban et al., 2021) and PHISTO (Durmuş Tekir et al., 2013) databases.
2.1.5 Annotations—structure, function and ontology
The sequences are mapped to the UniProt database to extract information on 3D-structure, functional attributes and ontology terms.
2.1.6 Kegg pathway mapping (pathogen vs. human)
It is important to identify drug targets that participate in pathogen specific pathways for minimum side effects. This sub-module identifies the metabolic pathways associated specifically with the pathogen proteins by mapping the sequences to KEGG database (Kanehisa et al., 2023).
2.2 Druggability analysis
The druggability of targets is predicted based on sequence similarity of pathogen proteins to experimentally validated druggable proteins of DrugBank 5.0 (Wishart et al., 2018), Therapeutic Target Database (TTD) (Zhou et al., 2023) and ChEMBL (Mendez et al., 2019) databases. This module also provides information on potential drugs or small molecules for these targets, based on the data available in these databases.
2.3 Immunoinformatics analysis
Effective immunization against infectious diseases is achieved through adaptive immunity that comprises of antigen-specific T cell and B cell mediated response. The sub-modules can be used to predict antigenic regions within a protein sequence as well as to identify B- cell and T-cell specific epitopes in the sequence.
2.3.1 Antigenicity prediction
This sub-module offers alignment-based as well as alignment-free methods for antigenicity prediction. The alignment-based method compares protein sequences with experimentally validated bacterial protective antigens derived from Protegen (Ong et al., 2017) database through BLASTp alignment scores. The alignment-free method uses Vaxijen 3.0 (Doytchinova & Flower, 2008) which transforms protein sequences into property-based vectors for antigen prediction. Users can opt for consensus based prediction of antigenic protein sequences from both methods.
2.3.2 B-cell epitope prediction
This sub-module is based on (i) Chou and Fasman Beta-Turn (Chou & Fasman, 1978), (ii) Emini Surface Accessibility (Emini et al., 1985), (iii) Karplus & Schulz Flexibility (Karplus & Schulz, 1985), (iv) Kolaskar & Tongaonkar (Kolaskar & Tongaonkar, 1990), and (v) Parker Hydrophilicity (Parker et al., 1986) based predictions. This module can be utilized to detect B-cell epitopes through a consensus prediction generated from various algorithms.
2.3.3 T-cell epitope prediction
This sub-module is based on IEDB developed algorithms NetMHCpan 4.1 (Parker et al., 1986) for MHC I and NetMHCIIpan 4.1 (Kaabinejadian et al., 2022) for MHC II binding predictions for available HLA alleles. Multiple sequences can be processed simultaneously to detect allele specific T-cell epitopes based on user defined peptide length.
The aforementioned modules and submodules can be linked through a hierarchical pipeline as per user specifications.
2.4 Systems biology analysis
Complex biological systems can be analyzed using systems biology tools to prioritize pathogen targets. This module has the following sub-modules; (1) topological network analysis, (2) essential metabolic reaction prediction, and (3) in silico gene knockout analysis. Topological network analysis can predict important nodes or proteins in a protein–protein interaction network based on degree and network centrality measures (Pinto et al., 2014). Essential metabolic reactions and critical enzymes of these pathways can also be predicted from pathogen's genome scale metabolic models using flux variability analysis and flux balance analysis respectively (Gu et al., 2019).
3 VALIDATION OF PBITV3
Our team had successfully verified the utility of PBITv1 and PBITv2 using the Candida proteome. About 45% of the PBIT predicted targets were documented in literature as essential proteins for Candida growth and pathogenicity. Further, in vitro assay using the drug predicted from the druggability module against YmL9 protein of Candida, was found to retard the pathogen's growth thereby authenticating the capability of the tool for prediction of novel drugs and targets (Mukherjee et al., 2021).
For validation of PBITv3, we have used Mycobacterium tuberculosis (Mtb), the causal organism for tuberculosis, and evaluated its antigenic proteins through PBITv3 workflow (Figure 2). Despite the availability of antibiotics, the search for novel targets and vaccine for TB continues due to the development and spread of resistance to current drugs. Since the proteome of Mtb is well characterized and researched for identifying drug targets and vaccine candidates, it was used for evaluating and validating the PBIT workflow. The goal of this exercise was not to identify novel targets or antigens, but rather to leverage reproducibility of previously published findings for validation of algorithm.
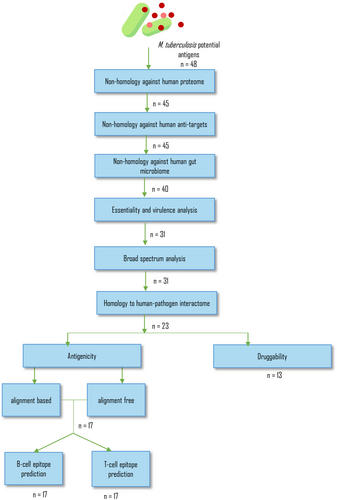
A dataset of 48 potential antigens of Mtb that included in vivo expressed (IVE)-TB antigens, latent antigens, hypoxia related proteins and conjugated protein subunit antigen Mtb72F sequence was compiled from literature (Coppola et al., 2021; Bertholet et al., 2008; Skeiky et al., 1999, 2004) (Table 2). These antigens have undergone experimental assessment to determine their capacity to induce an immunogenic response in murine host models. Hence, they serve as a suitable dataset for validating the application of the PBIT workflow in epitope identification for vaccine development. These 48 putative antigens were analyzed through specific modules of PBITv3 (Figure 2) as per the protocol adopted in publications (Sarom et al., 2018; Jalal et al., 2022) and the observations are discussed below.
| Sl no | Gene | Uniprot ID | PBITv3 modules | Literature evidences | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-homology to human proteome | Non-homology to anti-targets | Non-homology to gut-microbiota | Homology to essential or virulent proteins | Broad spectrum analysis | Homology to human–pathogen interactome | Antigenicity prediction | B-cell epitope (rank) | T-cell epitope (rank) | Druggability | ||||
| 1 | Rv0287/Rv0288 | O53692 | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | - | - | Probable association with drug resistance [PMID: 32379526]; potential vaccine candidate identified by MD simulation [PMID: 37079575] |
| 2 | Rv0440 | P9WPE7 | ✗ | - | - | - | - | - | - | - | - | - | Major immunoreactive essential protein; elicit robust proinflammatory responses from DCs and promote DC maturation and antigen presentation to T cells [PMID: 29133346] |
| 3 | Rv0470c | P9WPB3 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | ✔ | Drug target inhibited by Thiacetazone [PMID: 18094751]. Consistently recognized across mice both after Mtb challenge and produce significant cytokine response [PMID: 34083546] |
| 4 | Rv0642c | Q79FX8 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | ✔ | Drug target inhibited by thiacetazone [PMID: 18094751]. Non-significant cytokine production in mice tissue across time points [PMID: 34083546] |
| 5 | Rv0826 | O53837 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | Antigen recognized by T-cell [PMID: 34083546] |
| 6 | Rv0991 | O05574 | ✔ | ✔ | ✗ | - | - | - | - | - | - | - | Antigen recognized by T-cell [PMID: 34083546] |
| 7 | Rv1131 | I6Y9Q3 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 7 | 3 | ✔ | Antigen recognized by T-cell across multiple tissues and induced cytokine production [PMID: 34083546] |
| 8 | Rv1221 | P9WGG7 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | ✗ | Antigen induces TNF-α but not IFN-γ responses and recognized in few tissues and mice strains [PMID: 34083546]; essential gene for in vitro growth of H37Rv; associated with virulence in murine model [PMID: 36960291] |
| 9 | Rv1791 | Q79FK4 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 9 | 16 | ✗ | Antigen recognized by T-cell [PMID: 34083546] |
| 10 | Rv1846 | P9WMJ5 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | Antigen recognized by T-cell [PMID: 34083546] |
| 11 | Rv1872 | P9WND5 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | ✔ | Identified as drug-target [PMID: 19099550]. Low TNF-alpha & IL-17 response in d C3HeB/FeJ (C3H) mice [PMID: 34083546] |
| 12 | Rv1980c | P9WIN9 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | Predicted vaccine candidate from whole genome analysis [PMID: 18505592] co-expressing antigen of BCG recombinant DNA vaccine and efficacy studies in mice [PMID: 19284499, PMID: 21340709, PMID: 15498274] |
| 13 | Rv2461 | P9WPC5 | ✔ | ✔ | ✗ | - | - | - | - | - | - | - | Protein complex with ClpP2 and ClpC1 inhibited by antibiotics ecumicin and rufomycin [PMID: 36580851], antibiotic acyldepsipeptides (ADEP) dysregulate the Clp protease for unregulated proteolysis [PMID: 36286522] |
| 14 | Rv2626 | P9WJA3 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 16 | 13 | ✗ | Secretory functions. Strong humoral response in Balb/c mice [PMID: 17145953] |
| 15 | Rv2873 | P9WNF3 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 4 | 10 | ✔ | Cell surface lipoprotein Mpt83 (lipoprotein P23), stimulates antigen-specific T cell response [PMID: 22567094] |
| 16 | Rv3048c | P9WH71 | ✔ | ✔ | ✗ | - | - | - | - | - | - | - | Essential gene involved in the DNA replication pathway [PMID: 14573627] |
| 17 | Rv3052 | P9WIZ3 | ✔ | ✔ | ✗ | - | - | - | - | - | - | - | Essential gene for in vitro growth of H37Rv [PMID: 21980284] |
| 18 | Rv3583c | P9WJG3 | ✔ | ✔ | ✗ | - | - | - | - | - | - | - | Essential gene for in vitro growth of H37Rv [PMID: 21980284] |
| 19 | Rv3615 | P9WJD7 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 17 | 14 | ✗ | EspC contained broadly recognized CD4(+) and CD8(+) epitopes [PMID: 21427227] |
| 20 | Rv3616 | P9WJE1 | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | - | - | EspA, EspC and EspD form a complex and are MHC binding epitopes, induces TNF-α but not IFN-γ responses & recognized in few tissues & mice strains [PMID: 34083546] |
| 21 | Rv3846 | P9WGE7 | ✗ | - |
- | - | - | - | - | - | - | - | Superoxide dismutase, DNA vaccine expressing superoxide dismutase imparted maximum protection as observed by a 50 and 10 folds reduction in bacillary load [PMID: 16157425] |
| 22 | Rv3874/Rv3875 | P9WNK5 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 12 | 15 | ✗ | Epitope for fusion vaccine candidate [PMID: 31642227], DNA vaccine [PMID: 16157425], delayed-hypersensitivity [PMID: 10639479] |
| 23 | Rv1733c | P9WLS9 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | Synthetic long peptide derived from Rv1733c is well-recognized by T-cells [PMID: 26202436] |
| 24 | Rv2034 | O53478 | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | - | - | Transcriptional regulator, induces TNF-α but not IFN-γ responses [PMID: 34083546] |
| 25 | Rv3353c | O50382 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | IgG response to Rv2029c, Rv2031c, Rv2034, Rv2628, Rv3353c, ESAT6:CFP10, and chimeric PstS1 [PMID: 29523330] latency associated antigen [PMID: 26421415] |
| 26 | Rv2029c | P9WID3 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 6 | 7 | ✗ | Latency associated antigen [PMID: 26421415] |
| 27 | Rv1886c | P9WQP1 | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | - | - | Protein epitope a part of B21 DNA vaccine [PMID: 36569899], low IgG2c response [PMID: 34083546] |
| 28 | Rv1626 | P9WGM3 | ✔ | ✔ | ✗ | - | - | - | - | - | - | - | Two-component regulator pdtaR, higher IgG response to Rv1626 antigen on PHA beads [PMID: 28242005, PMID: 28714174] |
| 29 | Rv2875 | P9WNF5 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 10 | 12 | ✔ | Humoral response (IgG2c), predicted secreted protein—identified in culture filtrates of M. tuberculosis H37Rv, multistage antigen component of DNA-DMT vaccine [PMID: 29535714] |
| 30 | Rv3044 | O53291 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 13 | 5 | ✗ | Induces humoral response (IgG2c), multistage antigen component of DNA-DMT vaccine [PMID: 29535714] |
| 31 | Rv0496 | P9WHV5 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | ✔ | Identified as a drug target by deletion studies [PMID: 34728648]. Intermediate reduction in viable bacteria count after immunization [PMID: 19017986] |
| 32 | Rv0831c | O53842 | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | - | - | Serological marker conserved protein [PMID: 28223349] |
| 33 | Rv1813c | P9WLS1 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | Component of b21 DNA vaccine [PMID: 36569899] |
| 34 | Rv3020c | Q6MX18 | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | - | - | Immunodominant antigen in murine Mtb infection [PMID: 33240275] |
| 35 | Rv3619c | P0DOA7 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 14 | 17 | ✔ | Induces humoral response (IgG2c) and Th1 response [PMID: 32027660] |
| 36 | Rv0164 | L7N657 | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | - | - | Low immunogenicity and vaccine induced protection against Mtb in mice [PMID: 19017986] |
| 37 | Rv1590 | P9WLT7 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | Required for growth in C57BL/6J mouse spleen, by transposon site hybridization (TraSH) in H37Rv [PMID: 14569030] |
| 38 | Rv1818c | P9WIF5 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 15 | 2 | ✗ | B-cell humoral response [PMID: 17687113]; stimulated CD4+ and CD8+ T-cell proliferation as well as IFN-gamma secretion [PMID: 24904584] |
| 39 | Rv2032 | P9WIZ9 | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | - | - | Low immunogenicity and vaccine induced protection against Mtb in mice [PMID: 19017986] |
| 40 | Rv3620c | P9WNI3 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | Humoral response (IgG2c), Th1 response, [PMID: 31923726] |
| 41 | Rv2623 | P9WFD7 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 5 | 8 | ✗ | Induced by Th1 response [PMID: 12506197]; in-silico studies as potential drug target [PMID: 37878080, PMID: 25666036] |
| 42 | Rv2866 | O33348 | ✔ | ✔ | ✔ | ✗ | - | - | - | - | - | - | Overexpression inhibits mycobacterial growth in presence of human macrophages [PMID: 19114484] |
| 43 | Rv3029c | P9WNG7 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 8 | 9 | ✔ | Thiol specific oxidative response [PMID: 16006064], ethambutol targets [PMID: 29366429] |
| 44 | Rv3133c | P9WMF9 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 11 | 11 | ✔ | Transcription factor with role in dormancy regulation in latent tuberculosis [PMID: 18359816] |
| 45 | Rv3204 | O05862 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✗ | - | - | ✗ | Low immunogenicity [PMID: 19017986] |
| 46 | Rv0125 | O07175 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 3 | 6 | ✔ | C-terminal domain of Rv0125 (Mtb32C) can strongly motivate TCD8 cells, which produce cytokines [PMID: 15187142] |
| 47 | Rv1196 | L7N675 | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 2 | 4 | ✔ | Favors development of Th2-type response, and down-regulates the pro-inflammatory and Th1-type response [PMID: 19880448, PMID: 21451109]. |
| 48 | Mtb72F | CAR95102.1 (NCBI ID) | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | 1 | 1 | ✔ | Recombinant fusion proteins derived from Mtb32A and Mtb39A (encoded by Rv0125 and Rv1196, respectively) [PMID: 15187142] |
| Total number of proteins cleared the module | 46 | 46 | 40 | 31 | 31 | 23 | 17 | 13 | |||||
- Note: ✔ indicates proteins that cleared the module, ✗ indicates proteins that did not clear the module and was not considered for further analysis (indicated by -).
3.1 Screening and characterization
The proteins were screened for homology against human proteome, anti-target and gut microbiota to determine the specificity of the proteins to the pathogen. The threshold E-value and sequence identity were maintained at default values of 0.005 and 50% respectively. Following this, the proteins were analyzed for essentiality and virulence, broad spectrum activity and role in host–pathogen interaction with E-value and alignment length parameters set to 0.001 and 1% respectively.
3.1.1 Non-homology against human proteome, human anti-target, gut microbiota
Of the 48 sequences, 8 were screened out due to shared homology with either human proteome, anti-targets or gut microbiota. These eight proteins include heat-shock proteins, proteases and transcription factors that are evolutionary conserved amongst different organisms (Table 2).
3.1.2 Essentiality and virulence analysis
Of the remaining 40, 14 and 5 were homologous to known essential proteins and virulent proteins respectively. Twelve were homologous to both essential as well as virulent genes. Thirty-one proteins cleared this module; and of these, 14 have been experimentally validated as essential and virulent proteins (Appendix S2).
3.1.3 Broad spectrum analysis
All the 31 proteins were found to share significant similarity with proteomes of other pathogenic organisms and therefore could be classified as poly-microbial targets.
3.1.4 Homology to host–pathogen interactome
The involvement of the identified epitopes in host-pathogen interactions is crucial for vaccine development (Tsai et al., 2022). Of the 31 proteins, 23 were homologous to pathogen proteins that are known to interact with host (both human and non-human; see section 2.1). Eight antigens which were not homologous to the interactome were experimentally found to elicit low immunogenic or INF-γ response (Table 2).
3.2 Druggability analysis
Of the 23 proteins screened for druggability, 13 were found to be druggable based on their similarity to known drug targets. Five of the 13 proteins were found to be experimentally validated as targets of thiacetazone (Alahari et al., 2007) and ethambutol drugs (Ghiraldi-Lopes et al., 2019) and by gene deletion studies (Table 2).
3.3 Immunoinformatics analysis
3.3.1 Antigenicity prediction
Antigenicity of the 23 proteins were predicted by alignment free and alignment-based methods. Seventeen proteins, that were predicted to be antigenic by both methods, were screened for epitope prediction. It was observed that most of these six proteins which were predicted to have poor antigenicity exhibited low cytokine response in murine models, thus confirming the accuracy of PBITv3's antigenicity module (Table 2).
3.3.2 B-cell epitope prediction
Binding to B-cell epitope was predicted using all the five methods by maintaining a window size of six residues for each protein. B-cell epitopes were found in all the 17 proteins; these proteins were ranked based on the number of epitopes recognized by the algorithm (Table 2). It was observed that the synthetic vaccine construct Mtb72F ranked highest in terms of epitope sites. Some of the other high ranked proteins such as Mtb32A (Rv0125), Mtb39A (Rv1196), Mpt83 (Rv2873), Hrp1 (Rv2626), FecB (Rv3044) and PE-PGRS family protein (Rv1818c) have been investigated as important vaccine candidates. These results illustrate the module's efficacy in predicting vaccine candidates.
3.3.3 T-cell epitope prediction
T-cell binding epitope was predicted based on MHC-I and MHC-II alleles. For MHC-I, a minimum length of 8 residues was considered for binding to HLA-A*02:01 allele which is associated with vaccine efficacy (Gartland et al., 2014). MHC-II binding was evaluated keeping 11-mer peptide as window-size and DRB1*01:04 as the binding allele for its involvement with immune response to Mtb-antigens (Shams et al., 2004). All proteins were found to bind to MHC-I and MHC-II allele; higher number of epitopes were observed in Mtb72F protein vaccine. Abundant HLA binding epitopes were also detected for investigational vaccine candidates indicative of their immunogenicity (Table 2).
At the end of the workflow, PBITv3 successfully identified the most promising candidate Mtb72F fusion protein vaccine construct that has progressed through phase-II of clinical trial NCT01755598. It also identified additional drug targets and vaccine candidates that have either undergone experimental validation or are currently in the validation process. Through this example, the efficiency of PBITv3 as a rapid method for accurately predicting vaccine candidates and drug targets can be appreciated.
4 DEMONSTRATED UTILITY OF PBIT
Upon reviewing the citations of PBIT, it is gratifying to note that the tool that had been developed in 2016 and later updated in 2021, has been used globally by several research groups for drug target prediction and vaccine development. A synopsis of the cited utility is mentioned below.
4.1 Safety profile of multiepitope vaccine construct
- Sanches and team constructed a multi-epitope vaccine from Schistosoma mansoni and used PBIT to evaluate the safety of the vaccine through non-homology analysis (Sanches et al., 2021).
- Khalid et al. performed safety profiling of a multi-epitope vaccine construct from Borrelia burgdorferi through non-homology analysis to gut microbiota (Khalid et al., 2022).
- Nayak et al. adopted a reverse vaccinology approach to identify vaccine candidates from Mtb proteome. The team used PBIT to filter out proteins homologous to gut microbiota and selection of druggable proteins (Nayak et al., 2023).
- A vaccine construct against SARS-CoV2 was examined using PBIT screening modules (Gustiananda et al., 2021) for its safety profile.
- Gomes and co-workers used PBIT to assess homology between multi epitope chimeric protein and proteome of host and gut microbiota for developing vaccine against Treponema pallidum infection (Gomes et al., 2022).
- Non-homology analysis modules of PBIT were used to initially filter out proteins homologous to human proteome, anti-targets, and gut microbiota from pan-proteome of Mycobacteroides clade and subsequently to select essential and virulent proteins (Satyam et al., 2020).
4.2 Role in drug and vaccine target identification
- Cesur et al. used the druggability module of PBIT to identify druggable proteins of Klebsiella pneumoniae and later verified these targets for their presence in diverse infectious agents using broad spectrum analysis (Cesur et al., 2020).
- Canário Viana et al. identified four drugs and targets from the pan-proteome of 108 Corynebacterium strains using PBIT (Viana et al., 2022).
- Drug and vaccine targets were predicted from Bordetella pertussis and analyzed for essentiality and non-homology to gut microbiota using PBIT (Felice et al., 2022).
-
PBIT was used to screen out human and gut-microbiota homologs to identify putative targets from five Salmonella strains. Antigenic drug targets were further analyzed to predict vaccine candidates (Sah et al., 2020).
Similar studies were performed using PBIT to screen pathogen proteomes of Serratia marcescens (Prado et al., 2022), Pseudomonas aeruginosa (Rahman et al., 2023; Atron, 2023), Salmonella enterica serovar Typhimurium (Kocabaş et al., 2022), Rickettsia (Felice et al., 2022), Corynebacterium ulcerans, Corynebacterium silvaticum (Cerqueira et al., 2022) to identify potential drug and vaccine targets. In addition to high-throughput analysis, individual proteins, such as MEP2 protein in Candida albicans (Khalil, 2020), have also been assessed for their safety through the non-homology modules of PBIT.
4.3 Role in mRNA vaccine construction
mRNA-based vaccines contain the antigen gene flanked by 5′ and 3′ untranslated regions (UTRs), and additional nucleic acids required for mRNA stability. PBIT has been used to screen the translated mRNA sequences of final vaccine construct for pox viruses (Kovačić & Salihović, 2022) and Mtb (Kovačić et al., 2022) to verify the autoimmune potential (by comparing the similarity of epitopes to human proteins) and its effect on gut microbes. Although, a major drawback is the absence of wet-lab data in these studies, the mRNA vaccines have been computationally evaluated for their ability to elicit immune response and stability using MD simulation. Both the vaccine constructs were predicted to be antigenic, safe and efficacious.
Overall, these citations exemplify the contribution of PBIT in high-throughput identification of drug targets and design of vaccine candidates from pathogen proteomes.
5 CONCLUSIONS
Over the recent years, we have witnessed pandemics, epidemics and rise in drug-resistant infectious agents resulting in substantial morbidity and mortality across the globe. Emergence of new infectious diseases pose fresh challenges for therapeutic management strategies. The surge in availability of data related to pathogen proteomes and efficient database query algorithms makes it ideal to leverage computational methods to address the ever-growing demand for new drugs and targets. The development of PBIT has been our effort towards this goal. We found several citations for PBIT v1 and v2, wherein researchers have used the tool to identify potential pathogen targets for multiple pathogens such as P. aeruginosa, S. enterica, C. albicans and M. tuberculosis. In few cases, researchers had to rely on other algorithms for testing antigenicity or for epitope prediction of the proteins identified by PBIT. Therefore, in PBITv3, we have incorporated the module on immunoinformatics analysis, to facilitate accomplishment of the tasks related to identification of drug targets and vaccine epitopes within a single portal. Additionally, we have also integrated a systems biology based model to harness the power of metabolic pathway networks in target prediction. The updated and expanded version of PBIT will be a valuable tool for screening and prioritizing drug and vaccine candidates.
6 ADVANTAGES AND LIMITATIONS OF PBITV3
- To the best of our knowledge, PBIT is the only tool available online that can facilitate investigation of numerous established principles of target identification on a unified platform.
- Through the pipeline builder option, users can connect multiple modules, in their preferred order, seamlessly without the need to upload files at each step.
- Although several stand-alone servers are available to predict essentiality and virulence (DEG, VFDF), druggability (DrugBank, TTD) or antigenicity (Vaxijen, IEDB) of a protein sequence, they can be employed to test only one application/algorithm at a time. The strength of PBIT is its capacity to execute multiple applications and derive a consensus prediction from these algorithms.
6.1 Limitations
- The tool can process up to 500 sequences concurrently. Larger proteomes must be trimmed into multiple files for analysis.
- The immunoinformatics module has limited options for B-cell and T-cell prediction.
These limitations will be resolved in future updates.
AUTHOR CONTRIBUTIONS
Susan Idicula-Thomas: Conceptualization; methodology; funding acquisition; project administration; resources; writing – original draft; writing – review and editing; supervision; formal analysis; visualization. Shuvechha Chakraborty: Methodology; data curation; software; validation; writing – original draft; writing – review and editing; visualization; investigation. Mehdi Askari: Methodology; software; formal analysis; visualization; investigation. Ram Shankar Barai: Software; investigation; formal analysis; supervision; visualization.
ACKNOWLEDGMENTS
The authors are grateful to Dr. Geetanjali Sachdeva, Director, ICMR-NIRRCH for support. We thank Mr. Pankajkumar Pandey and Ms. Anam Arshi for technical assistance and Ms. Krisna Parab for assisting in review of literature.
FUNDING INFORMATION
This work was supported by research funds from Department of Biotechnology (DBT), India [BT/PR40165/BTIS/137/12/2021], Science and Engineering Research Board (SERB), India [CRG/2021/004937] and Senior Research Fellowship from Indian Council of Medical Research [Myco/Fell/14/2022-ECD-II].
CONFLICT OF INTEREST STATEMENT
None declared.