The role of MALAT1 and UCA1 long non-coding RNAs on the prognosis of patients with glioblastoma: A systematic review and meta-analysis
Abstract
Glioblastoma multiforme (GBM) is a common central nervous system malignancy with poor survival despite new treatments. Although some evidence demonstrated the prognostic effects of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and urothelial carcinoma associated 1 (UCA1) long non-coding RNAs (lncRNAs) in patients with GBM, a comprehensive study has not yet evaluated the clinical importance of these lncRNAs. Hence, this review aimed to predict the significance of expressions of MALAT and UCA1 lncRNAs in patients with GBM. Using proper keywords, a thorough literature search was performed via databases, including PubMed, Web of Knowledge, Scopus, and EMBASE until December 2024. The relationship between lncRNA expressions and overall survival (OS) in patients with GBM was assessed using hazard ratios (HR) and confidence intervals (95% CI), and the fixed and random effects models were used to estimate the pooled effect size. Also, the Newcastle-Ottawa Quality Assessment Scale was used as an appraisal tool. Among 1553 initially founded records, 13 studies were enrolled in the final analysis, consisting of 915 and 257 samples in the MALAT1 and UCA1 groups, respectively. Compared to the patients with low expression, those with high expression of MALAT1 had a mortality risk of 80% (HR = 1.8, 95% CI = [1.39, 2.33], p = .001). Additionally, the impact of UCA1 expression on patient prognosis indicated that lower OS among patients was correlated with high expression of UCA1; however, the meta-analysis was not performed for UCA1 due to a lack of adequate studies. According to our findings, high expression of MALAT1 was correlated with poor prognosis in patients with GBM.
1 INTRODUCTION
Glioblastoma multiforme (GBM) represents the most aggressive and common form of brain tumor.1 Due to the elevated mortality associated with GBM, the average overall survival (OS) for affected individuals is approximately 14 months.2 Consequently, numerous investigations have been undertaken to establish therapeutic strategies, encompassing pharmacological interventions and gene therapy, aimed at improving prognosis and extending OS.3 Furthermore, early detection and prompt treatment significantly influence patient outcomes; thus, alongside the advancement of therapeutic approaches, the development of diagnostic methodologies is equally critical.4
Long non-coding RNAs (lncRNAs) are transcripts from non-coding regions that are not at high risk for cancer. Through different mechanisms, these molecules lead to the progress and/or inhibition of tumorigenesis processes.5 LncRNAs include more than 200 nucleotides and, besides being non-protein-coding, have a crucial role in numerous gene expressions.6 Previous studies indicated the role of lncRNA in the pathogenesis and prognosis of cancers, for example, breast6 and gastric.7 Among all the lncRNAs, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and urothelial carcinoma associated 1 (UCA1) have a key role in the progression, response to treatment, and prognosis of cancers.8
Gokay et al. found that MALAT1 expression is an important factor in the prognosis of GBM patients.9 Also, Chen et al. demonstrated that increased MALAT1 expression leads to different responses to therapy in patients with GBM.10 According to Xin et al.'s study, down-regulation of UCA1 increases apoptosis and could change invasive behavior in glioma cells.11
Although Zhou et al.12 in 2018 provided a systematic review for evaluation of the role of lncRNAs on the prognosis of patients with glioma (both low- and high-grade), there is limited information regarding the role of UCA1 and MALAT1 on the prognosis of patients with glioma. Hence, this study aimed to investigate the prognostic impacts of UCA1 and MALAT1 lncRNA expressions among patients with GBM.
2 MATERIALS AND METHODS
This systematic review was performed according to the PRISMA 2020 checklist.13 The main question in this systematic review was “What is the effect of low- and high-expression of UCA1 and MALAT1 on the prognosis of patients with glioma?”
2.1 Eligibility criteria and study selection
This systematic study included studies based on the following criteria without considering language and time limitations. All observational studies (i.e., cohorts, case–control, and cross-sectional) and non-clinical trials that examined the expression of one of the target lncRNAs and reported overall prognosis of glioma patients, including OS (time from diagnosis to patient death) and progression-free survival (PFS, time from diagnosis to tumor recurrence), were included. Also, animal and cell-culture studies, as well as clinical trials, were excluded.
2.2 Search strategies
A systematic search was performed in various databases, including MEDLINE (via OVID SP), EMBASE, Scopus, and Web of Knowledge, using the following keywords: glioblastoma, GBM, glioma, glioblastoma multiform, prognosis, survival, OS, progression, progression-free survival, long-noncoding RNA, MALAT1, UCA1, and their Medical Subject Headings (MeSH) until December 2024. The full search strategies for the databases searched are listed in Data S1. To cover all studies, the reference lists of review articles and meta-analyses were manually checked, and abstracts of articles in conference proceedings published in the aforementioned databases were included in the study. Final articles were entered into EndNote version 20 software (Thomson Reuters, Philadelphia, PA, USA), and duplicates were removed by a researcher (MD). The titles and abstracts of identified studies were scrutinized for eligibility by four reviewers in two groups. The search was updated every 3 months until the article was published.
2.3 Data extractions
After selecting the relevant studies, full texts of the remaining articles were screened to extract the following data into a predesigned Excel form: first author name, year of publication, country, sample size, age, gender, KPS, WHO grade, type of sample, follow-up, and hazard ratio (HR) of MALAT1 or UCA1 expression. The data extraction process was performed independently by reviewers in two groups, and the final decision was reached by consensus.
2.4 Quality assessments
The Newcastle-Ottawa Quality Assessment Scale (NOQAS)14 was used by two reviewers independently in each group to evaluate the quality of the included studies. Using the NOQAS, the studies were evaluated in three domains (i.e., comparability, selection, and exposure) and rated, given a maximum of four, two, and four stars, respectively. The NOQAS items, score rates for each eligible study, and cut-off points for assigned quality (good, fair, and poor) are presented in Data S2.
2.5 Statistical analysis
All analyses were conducted using the Stata statistical package (StataCorp, 2021, Stata Statistical Software: Release 17. College Station, TX: StataCorp LLC). HRs with corresponding confidence intervals (CI) were extracted from the studies. Fixed and random effects models were used to estimate the pooled effect size. Chi-square test, as well as I and Tau squared statistics, were used to check for the heterogeneity among studies. A sensitivity analysis was also performed by applying the leave-one-out method to investigate whether the results were robust. Publication bias was evaluated by examining any obvious asymmetry within the funnel plot.
2.6 Ethics statement
This study was approved by the Research Ethics Committees of the Neuroscience Institute, Brain and Spinal Cord Injury Research Center (approval code: IR.TUMS.NI.REC.1401.022).
3 RESULTS
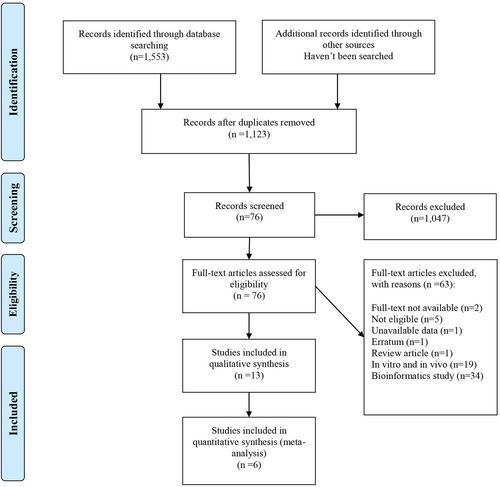
A total of 1553 records were identified through an initial database search, and after duplication removal, 1123 records were available for title screening. Following title and abstract screening, 76 articles were chosen for full-text evaluation, and finally, 13 studies10, 15-26 met our eligibility criteria for inclusion in the systematic review. Also, six studies10, 16, 18, 19, 24, 26 were available for the quantitative synthesis (Figure 1).
3.1 MALAT1
To review the role of MALAT1 lncRNA in patients' prognosis, nine studies10, 16, 18-21, 24-26 containing a total of 915 patients of various ages were included. Almost all studies had a minimum of 19 months of follow-up, and KPS was mentioned in only three studies,10, 24, 26 and all other effective health characteristics, such as gender, WHO grade, and sample (tissue or serum), are shown in Table 1. Fawzy et al.21 and Cao et al.26 suggested MALAT1 lncRNA as a promoter factor for OS, and other studies10, 16, 18-20, 24, 25 claimed that high expression of MALAT1 correlates with poor prognosis, in comparison with the low-expression group.
| Authors (ref) | Region | Age | Sample size | Sample | Follow up (months) | WHO grade | KPS | HR (95% CI) | Expression | OS (months) |
|---|---|---|---|---|---|---|---|---|---|---|
| Ma et al.18 | China | <45: 45 ≥45: 73 |
M: 63 F: 55 |
Tissue | 60 | I, II: 42 III, IV: 76 |
NM | Uni: 2.99 (1.82–4.93) Multi: 2.28 (1.372–3.81) |
High | Lower |
| Cao et al.26 | China | <60: 32 ≥60: 34 |
M: 36 F: 30 |
75 serum 66 tissue 141 total |
50 | I: 12 II: 9 III: 19 IV: 26 |
≥90: 19 <90: 47 |
Multi: 2.79 (1.15–4.73) in 66 tissue case | Low | Lower |
| Xiang et al.25 | China | NM | 37 | Tissue | NM | NM | NM | NM | NM | NM |
| Fawzy et al.21 | Egypt | Range: 35–60 | M: 28 F: 9 |
Tissue | 36< | IV: 37 | NM | NM | Low | Lower |
| Chen et al.10 | China | NM | 332 | Serum | 30 | IV: 332 | NM | Uni: 2.31 (1.2–4.87) Multi: 2.55 (1.22–5.2) |
High | NM |
| Shen et al.24 | United state | Mean: 58 Range: 45–70 |
M: 66 F: 40 |
Serum | Median:19 Range: 2–24 |
IV: 106 | 100 score: 23 90 score: 47 80 score: 19 80> score: 17 |
Multi: 0.94 (0.23–3.21) | High | Higher |
| Argadal et al.19 | Turkey | >45:51 <45: 24 |
M: 41 F: 34 |
Tissue | 60 | IV: 75 | NM | Multi: 2.12 (0.37–2.95) | High | Lower |
| Cheng et al.20 | China | <50:32 ≥50: 27 |
M: 36 F: 23 |
Tissue | 60 | I, II: 30 III, IV: 29 |
>80: 28 <80: 31 |
NM | High | Lower |
| Aksoy et al.16 | Turkey | 0–5:1 6–1–:3 11–18:5 19–45:24 46≤:52 |
M: 47 F: 38 |
Tissue | 19 | IV: 85 | NM | Multi: 0.65 (0.35–0.72) | High | Lower |
- Abbreviations: CI, confidence interval; F, female; HR, hazard ratio; KPS, Karnofsky performance status; M, male; NM, not mentioned; OS, overall survival.
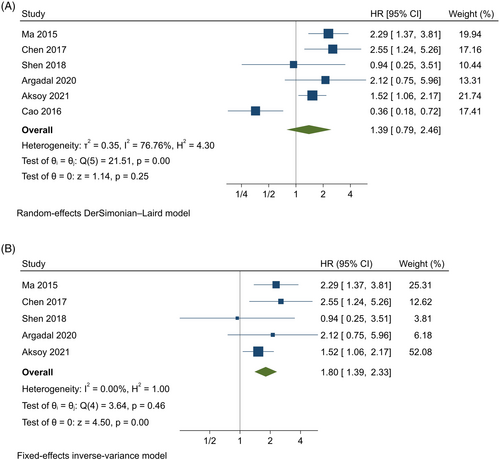
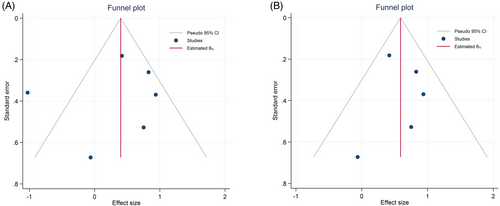
Six studies10, 16, 18, 19, 24, 26 were analyzed using the random-effects model with the DerSimonian–Laird method, indicating that high expression of MALAT1 was associated with a 40% increase in the risk of mortality compared to the low-expression group; however, it was not statistically significant (HR = 1.39, 95% CI = [0.78, 2.45], p = .253, Figure 2A). Besides, there was significant heterogeneity among studies (p < .001, I2 = 76.8%, T2 = 0.354) and also a marked asymmetry within the funnel plot (Figure 3A). Leave-one-out analysis revealed that the findings were sensitive to one26 of the studies. Therefore, the five studies10, 16, 18, 19, 24 were analyzed using a fixed-effects model with an inverse-variance method, indicating that high expression of MALAT1 was significantly associated with an 80% higher risk of mortality compared to the low expression group (HR = 1.8, 95% CI = [1.39, 2.33], p < .001, Figure 2B). Moreover, no significant heterogeneity was found among studies (p = .458, I2 = 0%), with no asymmetry within the funnel plot (Figure 3B).
3.2 UCA1
Regarding the effect of UCA1 lncRNA expression on patients' prognosis, four studies,15, 17, 22, 23 including 247 patients aged 25–55 years, were evaluated (Table 2). All studies had a minimum of 40 months of follow-up; the WHO grade was mentioned in only two studies,17, 22 and KPS was mentioned in only one study.22 Regarding Table 2, high expression of UCA1 lncRNA correlates with lower OS in patients. Although UCA1 was mentioned in the title and considered in the literature review, the meta-analysis could not be performed due to insufficient and inconsistent quantitative data across studies, which limited the ability to statistically synthesize its prognostic value. Since the HR was mentioned in only one study,22 we were unable to analyze its effect on prognosis quantitatively.
| Authors (ref) | Region | Age | Sample size | Sample | Follow up (months) | Who grade | KPS | HR (95% CI) | Expression | Os (months) |
|---|---|---|---|---|---|---|---|---|---|---|
| Zhao et al.22 | China | 40>: 25 40≤: 39 |
M: 35 F: 29 |
Tissue | 48 | I–II: 22 III–IV: 42 |
>80: 34 ≤80: 30 |
7.368 (3.032–17.9) | High | Lower |
| He et al.17 | China | <50: 55 ≥55: 25 |
M: 40 F: 40 |
Tissue | 36 | I–II: 27 III: 24 IV: 29 |
NM | 1.40 (0.60–3.27) | High | Lower |
| Sun et al.23 | China | NM | 63 | Tissue | 60 | NM | NM | NM | High | Lower |
| Li et al.15 | China | NM | 40 | Tissue | 60 | NM | NM | NM | High | Lower |
- Abbreviations: CI, confidence interval; F, female; HR, hazard ratio; KPS, Karnofsky performance status; M, male; NM: not mentioned; OS, overall survival.
3.3 Quality assessment
Regarding the NOQAS, four studies15, 20, 23, 25 were rated as poor quality (Table 3). Hence, the six studies10, 16, 18, 19, 24, 26 that were included in the quantitative synthesis had good quality. The NOQAS for all the studies is presented in Table 3.
| Studies (ref) | Selection | Comparability | Exposure | Quality | |||||
|---|---|---|---|---|---|---|---|---|---|
| Adequate definition of patient cases | Representativeness of patient case | Selection of controls | Definition of controls | Comparability of cases and controls | Ascertainment of exposure | The same method of ascertainment for participants | Nonresponse rate | ||
| Ma et al.18 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 8 |
| Xiang et al.25 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 5 |
| Cao et al.26 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 0 | 7 |
| Chen et al.10 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 0 | 7 |
| Fawzy et al.21 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 0 | 7 |
| Shen et al.24 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 9 |
| Argadal et al.19 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 8 |
| Cheng et al.20 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 5 |
| Aksoy et al.16 | 1 | 1 | 0 | 1 | 2 | 1 | 1 | 0 | 7 |
| Zhao et al.22 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 6 |
| He et al.17 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 6 |
| Sun et al.23 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 5 |
| Li et al.15 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 5 |
- Abbreviations: ■, poor; ■, fair; ■, good.
4 DISCUSSION
Numerous lncRNAs such as HOTAIR, MALAT1, H19, and UCA1 have been reported as important regulators in various aspects of tumorigenesis, referring to proliferation, migration, invasion, and metastasis.27 Indeed, the significant role of lncRNAs in the development and progression of various tumors has drawn attention to these RNA molecules.
4.1 UCA1
In our study, it was found that UCA1 is up-regulated in glioma tissues in comparison with normal brain tissues,15, 17, 22, 23 leading to a poor prognosis in glioma patients.17, 22
Previous studies have shown that UCA1—a noncoding bladder cancer-specific lncRNA—is abnormally expressed in various malignancies, including bladder, gastric, tongue squamous cell carcinoma, melanoma, esophageal, hepatic, colorectal, pancreatic, breast, and ovarian.22, 28
The regulatory function of UCA1 in cellular pathways was observed in preclinical studies. There was a negative correlation between UCA1 expression and miR-135a levels in tumor proliferation and migration in glioma.15, 29 Moreover, it was found that miR-182 overexpression inhibited the expression of UCA1, which led to reduced cell proliferation of glioma.17 In addition, a negative correlation between UCA1 and miR-122 was also observed.30 Indeed, miR-122 expression could act as a tumor suppressor by inhibiting proliferation and inducing apoptosis.30 So the interaction between UCA1 and miRNAs plays an important role in the development of glioma.
Also, UCA1 up-regulation can activate HOXD9, resulting in decreased cell proliferation, cell cycle arrest, and the induction of apoptosis in the U87 glioma cell line.15, 31
Regarding the clinicopathological features, He et al.17 reported that UCA1 was significantly related to both tumor size and grade, while Zhao et al.22 found no correlation between UCA1 expression and tumor size. Different cut-off values for UCA1 may influence the correlation between UCA1 expression and tumor size.
Overall, UCA1 is involved in the proliferation, apoptosis, migration, and invasion of tumor cells by regulating various cellular pathways, suggesting that UCA1 functions as an oncogene in brain glioma. Hence, UCA1 can be used as a prognostic predictor of glioma.
4.2 MALAT1
Our analysis has proven that up-regulation of MALAT1 is associated with lower OS.10, 16, 18, 19, 24, 26 Of note, we performed the meta-analysis twice, once with Cao et al.26 and once without, due to its contradictory result with other included studies in the quantitative synthesis.
The lncRNA MALAT1 is a highly conserved nuclear ncRNA18 and has an important role in normal brain functions, including cell proliferation, invasion, and metastasis in cancer.32 Dysregulation of MALAT1 is associated with metastasis and the risk of tumor recurrence after treatment in various cancer types, including myeloma, esophageal squamous cell carcinoma, colorectal, and pancreatic cancers, etc.10, 33 Nevertheless, previous studies reported contradictory results on the expression profile of MALAT1 and its correlation with prognosis in glioma.
A systematic review by Tian et al.33 found that MALAT1 expression was increased in various types of cancer, and it was significantly associated with poor OS. Similarly, Zhou et al.12 systematically assessed the expression of MALAT1 and other lncRNAs on patient survival and found that high MALAT1 expression was statistically associated with a poor prognosis. Xiang et al.25 found that the expression level of MALAT1 was much higher in glioma tissues than in para-cancerous tissues. The overexpression of MALAT1 in tumor cells compared to other normal tissues and its correlation with lower OS are supported by more studies.16, 18, 19, 24, 34 In contrast, Fawzy et al.21 demonstrated that the MALAT1 level was reduced in Egyptian individuals with GBM. They found that decreased MALAT1 expression was associated with tumor recurrence, reduced OS, and shorter disease-free survival (DFS).
Also, in vitro findings demonstrated that the expression level of MALAT1 is higher in tumoral cell lines in comparison to normal cell lines, and suppression of MALAT1 induced cell cycle retardation, cell apoptosis, and reduced the invasiveness of glioma cells.20, 35, 36 In contrast, Han et al.37 reported that MALAT1 acts as a tumor suppressor via down-regulation of MMP2 and inactivation of ERK/MAPK signaling. Similarly, the tumor-suppressive role of MALAT1 is further demonstrated by a novel regulatory pathway in which it suppresses cell viability by down-regulating miR-155 and promoting FBXW7 expression.26
Furthermore, previous studies had investigated the correlation between clinicopathological features and increased expression of MALAT1, including the association of MALAT1 expression with factors such as gender, age, tumor location, and tumor recurrence rate. It was shown that dysregulation of MALAT1 was associated with younger age, left-sided GB localization,19 tumor size, WHO grade,18, 20 and KPS.26 However, some studies showed the opposite results. It was shown that elevated levels of MALAT1 were associated with larger tumor sizes.18 In contrast, Cao et al. proposed that decreased MALAT1 expression was associated with larger tumor sizes. This discrepancy could be attributed to the different evaluation criteria used for measuring both MALAT1 expression and tumor size.26 Thus, MALAT1 may potentially be used as a new prognostic marker to predict the OS in patients with glioma.
4.3 Study limitations
Several limitations in this systematic review and meta-analysis should be emphasized. First, the criteria for calculating the cut-off value were not the same in different studies. Second, in our study, meta-analysis has not been performed on UCA1 due to insufficient statistical data. Third, the overall results seem to be affected by the low quality of included studies according to the NOQAS, especially in the UCA1 group.
4.4 Recommendations
Overall, based on this systematic study, further large-sized and good-quality studies are required to verify our results and confirm the clinical value of UCA1 and MALAT1 expression in glioma patients.
In summary, our study demonstrates that MALAT1's expression in glioma tissue can be either up- or down-regulated, and UCA1 is up-regulated in tumoral tissue compared to non-cancerous tissue, and its prognostic impact appears to be predominantly linked with its overexpression, which is associated with poorer patient outcomes and lower OS.
AUTHOR CONTRIBUTIONS
Sedighe Hooshmandi: Conceptualization; methodology; formal analysis; investigation; validation; writing—original draft; writing—review and editing. Ehsan Jangholi: Conceptualization; methodology; formal analysis; investigation; validation; writing—original draft; writing—review and editing. Amirreza Heidarian: Investigation; validation; writing—original draft; writing—review and editing. Mahsa Dehghani: Methodology; writing—review and editing; writing—original draft; validation; data curation. Aida Heidary: Methodology; data curation; validation; writing—review and editing; writing—original draft. Ali Ghassemi: Writing—review and editing; writing—original draft; validation; data curation; methodology. Ali Rezvanimehr: Writing—original draft; writing—review and editing; validation; data curation. Seyed Mohammad Ghodsi: Writing—review and editing; writing—original draft; methodology; conceptualization; validation; investigation. Mohammad Hoseinian: Writing—review and editing; writing—original draft; methodology; validation. Erfan Sadeghi: Writing—review and editing; writing—original draft; validation; methodology; investigation; formal analysis. Mostafa Farzin: Writing—review and editing; writing—original draft; methodology; validation. Hamid Zaferani Arani: Writing—review and editing; writing—original draft; methodology; validation. Mahmoudreza Hadjighassem: Writing—review and editing; writing—original draft; validation; methodology; conceptualization; supervision.
ACKNOWLEDGMENTS
We thank the Brain and Spinal Cord Injury Research Center, Neuroscience Institute (Tehran University of Medical Sciences) for providing financial support for the current research (grant number: 57826).
CONFLICT OF INTEREST STATEMENT
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




