Quantitative Proteomics Analysis Revealed the Potential Role of lncRNA Ftx in Promoting Gastric Cancer Progression
Abstract
Purpose
The molecular pathogenesis of gastric cancer is still ambiguous till now. Here, it is demonstrated that long noncoding RNA (lncRNA) Ftx acts as a novel tumor promotor of gastric cancer and a potent regulator of hexokinase-2 (HK2).
Experimental design
The role of lncRNA Ftx is detected in the loss and gain-of-function models of gastric cancer cells. Tandem mass tags combined with multidimensional LC and MS analyses are performed to decipher comparative proteomic profiles of gastric cancer cells in response to lncRNA Ftx knockdown and overexpression. Real-time roteomics-clinical applications (PCR) and western blot are used to validate the proteomic data.
Results
A total of 5124 proteins are quantified and indicated in diverse biological functions and metabolic related signaling pathways. Interestingly, HK2, which is downregulated when lncRNA Ftx is deleted and upregulated while lncRNA Ftx is overexpressed, is further validated in gastric cancer cells.
Conclusions and clinical relevance
The present study suggests lncRNA Ftx promotes gastric cancer progression by upregulating HK2, which provides a new perspective for the mechanism of gastric cancer progression, and thus identifies potential therapeutic targets for gastric cancer.
1 Introduction
Gastric cancer is one of the most frequent malignant tumors due to its undetectable onset and rapid progression, which contributes gastric cancer to be the third leading cause of cancer mortality worldwide.1 The occurrence of gastric cancer is associated with many risk factors, including a variable combination of chronic Helicobacter pylori infection, Epstein–Barr virus infection, other environmental factors, and host characteristics.2 Though the pathogenesis of gastric cancer has currently been studied extensively, the mysterious veil on this disease is still not fully uncovered. Therefore, revealing the cellular and molecular pathogenesis and finding key molecules involved in the process will be critical for upgrading therapeutic targets and treatment strategies.
Long noncoding RNAs (lncRNAs, defined as RNA > 200 nucleotides in length) have emerged as multifunctional regulators in diverse physiological progress and a myriad of diseases.3 lncRNA Ftx is a highly conserved transcript of 2300 nucleotides encoded by Ftx, which is an evolutionarily conserved noncoding gene that is located within the X-inactivation center (Xic).4 Ftx affects Xist expression and chromatin structure within the Xic region, and acts as an essential positive regulator of X chromosome inactivation (XCI).4 Yusuke et al. found that female mice lacking Ftx lncRNA exhibit impaired XCI and a microphthalmia-like phenotype.5 Furthermore, it is reported that lncRNA Ftx plays significant roles in regulating cardiomyocyte apoptosis, inhibiting hippocampal neuron apoptosis, abnormal activation of macrophage in cirrhosis as well as a variety of cancers including colorectal cancer, hepatocellular carcinoma, gliomas, and renal cell carcinoma.6-9 However, whether lncRNA Ftx plays a role in the oncogenic process of gastric cancer remains obscure.
The present study evaluated the potential role of lncRNA Ftx in gastric cancer cells and thus demonstrated lncRNA Ftx significantly promoted the progression of gastric cancer. Taking advantage of tandem mass tags (TMT) combined with multidimensional LC and MS analyses, the comprehensive proteomic profiles in response to lncRNA Ftx knockdown and overexpression in gastric cancer cells were performed to investigate the mechanism of lncRNA Ftx in exerting its oncogenic role in gastric cancer. Through quantitative proteomics, biochemistry assay, and bioinformatic analysis, differentially quantified proteins identified from the altered proteome were characterized. Moreover, through deep excavation of the quantified proteomics data, we found that the abundance of hexokinase-2 (HK2) was reduced when lncRNA Ftx was successfully deleted and increased while lncRNA Ftx was overexpressed. Further validation was carried out in gastric cancer cells by real-time PCR and western blot, and the results proved that lncRNA Ftx could significantly upregulate both the transcriptional and translational levels of HK2. Thus, we speculated that lncRNA Ftx promoted gastric cancer progression by positive regulation of HK2, and this provided a new insight into the pathogenesis of gastric cancer and highlighted promising therapeutic targets, which could contribute to improvement of approaches in gastric cancer therapy.
2 Results
2.1 lncRNA Ftx Promoted Malignant Behaviors of Gastric Cancer Cells
To investigate whether lncRNA Ftx plays a role in the progression of gastric cancer, we detected the function of lncRNA Ftx in MGC80-3 cells. We constructed a loss-of-function model by transfection of lentivirus-mediated lncRNA Ftx-RNA interference (Sh-Ftx) into MGC80-3 cells, while MGC80-3 cells transfected with Sh-NC were used as its mock control. A gain-of-function model was generated by lentivirus-mediated lncRNA Ftx overexpression (Ftx) in MGC80-3 cells, and MGC80-3 cells transfected with Ftx-NC were used as its mock control. The successful knockdown of lncRNA Ftx was confirmed by real-time PCR (Figure 1a). Our data showed that the proliferation (Figure 1b), invasion (Figure 1c), and colony formation (Figure 1d) of these lncRNA Ftx-deleted cells were significantly suppressed.
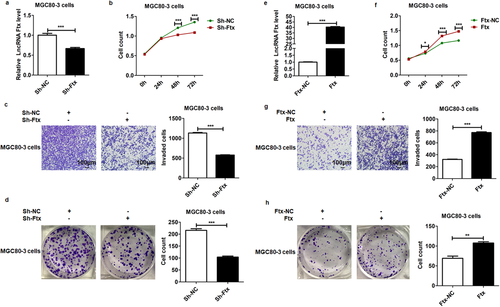
In contrast, after successful overexpression of lncRNA Ftx in MGC80-3 cells (Figure 1e), the proliferation (Figure 1f), invasion (Figure 1g), and colony formation (Figure 1h) of these lncRNA Ftx-overexpressed cells were all significantly promoted. Altogether, our data showed that lncRNA Ftx acted as a tumor promotor and facilitated the malignant behaviors of gastric cancer cells.
Statement of Clinical Relevance
This study demonstrated that lncRNA Ftx acted as a novel tumor promotor in progression of gastric cancer. The proteomic analysis provided the first global data of comprehensive changes in the quantitative proteome profiles in gastric cancer cells in response to lncRNA Ftx knockdown and overexpression. Further excavation and validation revealed HK2 was regulated by lncRNA Ftx. Taken together, this study identified a novel regulation mode of HK2 by lncRNA Ftx in the progression of gastric cancer and thus provided promising drug targets to achieve improvement in therapeutic strategy of gastric cancer.
2.2 Profiles of Proteome in the Loss and Gain-of-Function Models of MGC80-3 Cells
To further elucidate the molecular mechanism of lncRNA Ftx in regulating gastric cancer oncogenesis, we performed quantitative proteomics to profile the differentially expressed proteins in the loss and gain-of-function models of MGC80-3 cells, respectively, and further defined the crucial molecules and pathways involved in the carcinogenesis process of lncRNA Ftx. A total of 5784 proteins were identified by high-resolution LC–MS/MS analysis, among which 5124 proteins were quantified. Quantifiable proteins with quantification ratio of >1.3 were considered as upregulated, whereas those <0.769 were regarded as downregulated. All identified quantifiable proteins with increased (>1.3-fold) or decreased (<0.769) expression levels are listed in Supporting Information 1 and 2 (p < 0.05).
Compared with MGC80-3 cells transfected with Sh-NC, knockdown of lncRNA Ftx induced 229 differentially expressed proteins, including 152 upregulated proteins and 77 downregulated proteins. In parallel, overexpression of lncRNA Ftx led to 71 differentially expressed proteins compared with MGC80-3 cells transfected with Ftx-NC, which were composed of 28 upregulated proteins and 43 downregulated proteins. Therefore, these data indicated that lncRNA Ftx did, in fact, arouse alterations in the proteomic profiles of MGC80-3 cells and that variation might subsequently result in change of the on–off switch in the carcinogenesis process of gastric cancer.
2.3 Functional Annotation and Categories of Differentially Quantified Proteins
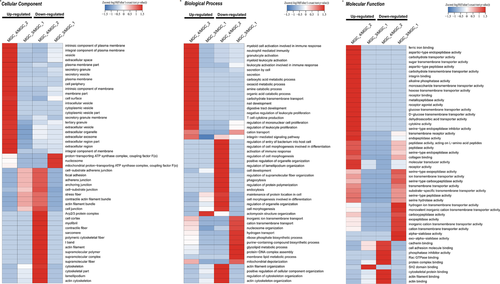
To screen the related biological function classification of differentially expressed proteins induced by lncRNA Ftx knockdown or overexpression in MGC80-3 cells, we analyzed the quantifiable proteome data through GO analysis. Hence, those differentially expressed proteins were annotated and classified into three categories: biological process, cellular component, and molecular function. The altered biological functions and the number of related proteins in lncRNA Ftx knockdown or overexpression group were analyzed and compared. The top five affected biological functions and the amount of differentially expressed proteins are summarized in Table 1. Moreover, the subcellular location analysis indicated that the differentially quantified proteins were wildly distributed in subcellular structures (Figure S1, Supporting Information 3). Taken together, our GO analysis suggested that the differentially quantified proteins were associated with a myriad of biological processes including cellular and metabolic process, involved in varieties of cellular components, and have a wide range of molecular functions in MGC80-3 cells.
| A | ||||
|---|---|---|---|---|
| lncRNA Ftx knockdown | Up classification | No. proteins [%] | Down classification | No. proteins [%] |
| Biological process | Cellular process | 134 (15) | Cellular process | 72 (15) |
| Single-organism process | 125 (14) | Single-organism process | 59 (13) | |
| Metabolic process | 98 (11) | Biological regulation | 58 (12) | |
| Biological regulation | 89 (10) | Cellular component organization or biogenesis | 44 (9) | |
| Response to stimulus | 84 (10) | Response to stimulus | 37 (8) | |
| Cellular component | Cell | 147 (24) | Cell | 77 (24) |
| Organelle | 138 (23) | Organelle | 72 (23) | |
| Membrane | 99 (16) | Membrane | 36 (11) | |
| Extracellular region | 82 (14) | Extracellular region | 34 (11) | |
| Membrane-enclosed lumen | 59 (10) | Macromolecular complex | 33 (10) | |
| Molecular function | Binding | 133 (49) | Binding | 73 (59) |
| Catalytic activity | 69 (26) | Catalytic activity | 21 (17) | |
| Transporter activity | 15 (6) | Molecular function regulator | 10 (8) | |
| Molecular function regulator | 14 (5) | Structural molecule activity | 8 (6) | |
| Molecular transducer activity | 12 (4) | Signal transducer activity | 5 (4) | |
| B | ||||
|---|---|---|---|---|
| lncRNA Ftx overexpression | Up classification | No. Proteins [%] | Down classification | No. Proteins [%] |
| Biological process | Cellular process | 25 (13) | Cellular process | 39 (16) |
| Single-organism process | 24 (13) | Metabolic process | 36 (15) | |
| Biological regulation | 22 (12) | Single-organism process | 34 (14) | |
| Cellular component organization or biogenesis | 18 (10) | Biological regulation | 24 (10) | |
| Developmental process | 15 (8) | Response to stimulus | 20 (8) | |
| Cellular component | Cell | 26 (27) | Cell | 42 (24) |
| Organelle | 20 (21) | Organelle | 37 (21) | |
| Membrane | 13 (14) | Membrane | 26 (15) | |
| Membrane-enclosed lumen | 9 (10) | Extracellular region | 21 (12) | |
| Cell junction | 8 (9) | Macromolecular complex | 20 (12) | |
| Molecular function | Binding | 28 (64) | Binding | 34 (46) |
| Catalytic activity | 10 (23) | Catalytic activity | 19 (26) | |
| Molecular transducer activity | 2 (5) | Transporter activity | 7 (9) | |
| Molecular function regulator | 1 (2) | Molecular function regulator | 4 (5) | |
| Signal transducer activity | 1 (2) | Signal transducer activity | 3 (4) | |
2.4 Functional Enrichment and Clustering Analysis of Differentially Quantified Proteins
The enrichment and clustering analyses were then carried out to identify the nature of differentially expressed proteins on account of lncRNA Ftx knockdown or overexpression (Figure 2; Figure S2, Supporting Information 3). First, we performed the clustering analysis based on the GO enrichment. As for the cellular component, the upregulated components including membrane part and intracellular vesicle were significantly enriched due to lncRNA Ftx knockdown, whereas cytoskeleton and actin filament were enriched with downregulated proteins (Figure 2a). On account of lncRNA Ftx overexpression, proteins related to actin filament bundle and stress fiber were enriched in the upregulated proteins, while proteins related to mitochondrial proton and nucleosome were enriched in the downregulated proteins (Figure 2a). In the biological process, upon knockdown of lncRNA Ftx, upregulated proteins were related to oxoacid metabolic process and amine catabolic process, whereas the downregulated proteins were related to activation of immune response and cell development (Figure 2b). Meanwhile, while lncRNA Ftx was overexpressed, the upregulated proteins appeared to be related to biological process including actomyosin structure organization, and the downregulated proteins were related to ribose phosphate biosynthetic process and glycolipid metabolic process (Figure 2b). On the ontology of molecular function, as lncRNA Ftx knockdown, proteins related to alkaline phosphatase activity and receptor binding were enriched in the upregulated proteins, and proteins related to cadherin binding and cytoskeletal protein binding were enriched in the downregulated proteins (Figure 2c). While lncRNA Ftx was overexpressed, proteins related to SH2 domain binding were enriched in the upregulated proteins, and proteins related to carboxypeptidase were enriched in the downregulated proteins (Figure 2c).
Then, we further performed a pathway enrichment-based clustering analysis of the lncRNA Ftx-responsive proteome according to the pathways identified by KEGG analysis. It appeared that proteins expression involved in peroxisome and fatty acid degradation were upregulated pathways after knockdown of lncRNA, while galactose metabolism, hippo signaling pathway, and adenosine 5′-monophosphate-activated protein kinase (AMPK) signaling pathway were downregulated pathways (Figure 3a). Meanwhile, overexpression of lncRNA Ftx led to upregulated pathways including p53 signaling pathway, and downregulated pathways such as oxidative phosphorylation and N-glycan biosynthesis (Figure 3a).
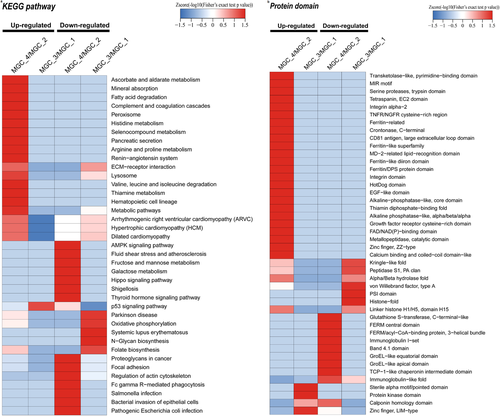
To define the domain characteristics of the differentially quantified proteins induced by lncRNA Ftx knockdown and overexpression, we further carried out the analysis of the domain enrichment on the basis of the previous domain analysis. The results implicated that the upregulated proteins affected by lncRNA Ftx knockdown contained zinc finger (ZZ-type) and calcium binding and coiled-coil domain-like domain, while downregulated proteins contained FERM central domain and band 4.1 domain (Figure 3b). After overexpression of lncRNA Ftx, the upregulated proteins contained protein kinase domain and zinc finger (LIM-type), whereas downregulated proteins contained histone-fold and PSI domain (Figure 3b).
2.5 Validation of HK2 in MGC80-3 Cells as a lncRNA Ftx Regulated Protein
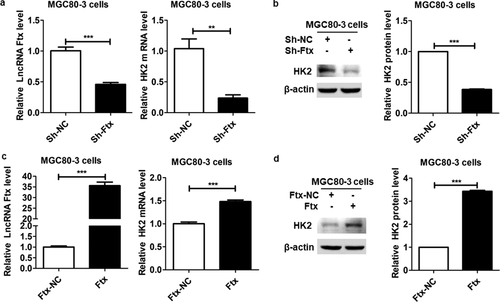
According to the proteome quantitative results, in order to focus on key proteins that were responsible for the oncogenic role of lncRNA Ftx, we paid great attention to the proteins with significant expression changes both responding to knockdown and overexpression of lncRNA Ftx, for example, HK2, the fold change of which was 0.673 and 1.378 upon lncRNA Ftx knockdown and overexpression, respectively. The abundance of HK2 on transcriptional and translational levels were verified using real-time PCR and western blot in MGC80-3 cells. Our data showed that knockdown of lncRNA Ftx dramatically reduced the mRNA and protein abundance of HK2 (Figure 4a,b). On the contrary, overexpression of lncRNA Ftx significantly increased the mRNA and protein levels of HK2 (Figure 4c,d). Altogether, these data indicated that lncRNA Ftx could regulate the abundance of HK2, and probably exerted its oncogenic effect by positively regulating the expression of HK2.
3 Discussion
The dysregulation of lncRNAs is considered as a pivotal driving force in tumorigenesis of various cancers including gastric cancer.10 Emerging evidence demonstrated that lncRNAs with upregulated or downregulated levels were involved in tumor growth, proliferation, apoptosis, cell cycle, invasion, and metastasis of gastric cancer.11
Recent researches have reported that lncRNA Ftx plays vital roles in tumorigenesis and progression of human malignancies. lncRNA Ftx promotes colorectal cancer progression by sponging miR-215 and inhibiting phosphorylation of vimentin.7 In hepatocellular carcinoma, lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPAR-γ pathway.8 Besides, lncRNA Ftx was proved to be associated with gliomas, and contribute to the development of drug resistance in acute myeloid leukemia.9, 12
In this study, we verified that lncRNA Ftx dramatically promoted the malignant behaviors of gastric cancer cells, such as proliferation, invasion, and colony formation. To understand the potential cellular and molecular mechanisms involved in this process, we systematically characterized the global proteome profiles of gastric cancer cells in response to lncRNA Ftx deletion and overexpression by taking advantage of quantitative proteomics, biochemistry analysis, and bioinformatic analysis. Currently, quantitative proteomics analysis is well developed and widely used in defining quantifiable proteins involved in diverse biological processes, cellular components, molecular functions, subcellular locations, signaling pathways, as well as protein domains. Furthermore, through deep excavation of quantitative proteomics data, we focused our sight on a differentially expressed protein, HK2, which was downregulated in response to lncRNA Ftx knockdown and upregulated while lncRNA Ftx was overexpressed. Further real-time PCR and western blot assays in gastric cancer cells validated the results of quantitative proteomics.
HK2 is one of the four major hexokinase isoforms, which catalyze the first committed step of glucose metabolism, phosphorylating glucose to glucose 6-phosphate via phosphate transfer from ATP.13 Accumulating evidence implicates that HK2 plays essential roles not only in glycolysis but also in cell survival via controlling cellular growth, preventing mitochondrial death and enhancing autophagy.14 In addition, HK2 is also extremely upregulated in many different types of human malignancies associated with enhanced aerobic glycolysis, the Warburg effect, which is responsible for the accelerated glucose metabolism in cancer cells and distinguishes them from normal cells.15 In rapidly proliferating tumor cells, to provide sufficient metabolic intermediates including nucleic acid, lipid, and protein synthesis to satisfy anabolic processes, a high rate of glucose metabolism under aerobic conditions followed by lactic acid fermentation make up for 60% of ATP consumption and HK2 stands out as the major contributor in this process.16 HK2 bound to the outer mitochondrial membrane via the porin-like protein voltage dependent anion channels, directly facilitating cancer cells growth and immortalization, thus maintaining the highly malignant state and increasing the possibility of cancer metastasis.17 Furthermore, HK2 overexpression is closely related to poor prognosis in solid tumors of digestive system such as gastric cancer, hepatocellular carcinoma, and colorectal cancer.18
Several previous studies have shown that a broad range of systemic and cellular mechanisms facilitate the transcriptional and translational levels of HK2 in different malignant cancers including epigenetic events such as demethylation, gene amplification, transcriptional, and post-transcriptional events.19 In the present study, our data suggested that lncRNA Ftx could significantly promote the mRNA and protein levels of HK2, which might be the reason why lncRNA Ftx exerted a critical oncogenic role in the progression of gastric cancer.
In conclusion, we for the first time demonstrated that lncRNA Ftx acted as a novel tumor promotor in the progression of gastric cancer. Combining with quantitative proteomics, biochemistry assay, and bioinformatic analysis, this study provided the first global data of comprehensive changes in the quantitative proteome profiles in gastric cancer cells in response to lncRNA Ftx knockdown and overexpression. Protein annotation, enrichment, and clustering analysis revealed the nature of the differentially quantified proteins identified from the quantitative proteomic data. Further excavation and validation revealed HK2 was regulated by lncRNA Ftx in gastric cancer cells. Taken together, this study identified a novel modulation mode of HK2 by lncRNA Ftx in the oncogenesis of gastric cancer and thus provided promising drug targets to achieve improvement in the therapeutic strategy of gastric cancer.
4 Experimental Section
Cell Culture and Transfection
Human MGC80-3 cells were obtained from the Cell Bank of Chinese Academy of Science (Shanghai, China) and cultured in DMEM (Hyclone, Massachusetts, USA) supplemented with 10% FBS, at 37 ˚C in a humidified atmosphere containing 5% CO2.
Construction of Genetically Modified Stable Cell Lines
Transfection of Sh-Ftx or Sh-NC and Ftx or Ftx-NC was performed using lentivirus (Shanghai GeneChem, Shanghai, China), according to the manufacturer's protocol. To produce cell lines with stably interfered expression of lncRNA Ftx, MGC80-3 cells were transfected with Sh-Ftx and selected with puromycin (2 µg mL–1), and MGC80-3 cells stably transfected with Sh-NC were used as its mock control. To obtain cell lines stably overexpressing lncRNA Ftx, MGC80-3 cells were transfected with Ftx and selected with puromycin (2 µg mL–1), while MGC80-3 cells stably transfected with Ftx-NC were used as its mock control. The stable lncRNA Ftx-interfered or overexpressing cell lines were validated by real-time PCR.
Real-Time PCR Assay
Total RNA was extracted from MGC80-3 cells, and real-time PCR was performed as described before.20 Primers for the human lncRNA Ftx were forward: 5′-GAATGTCCTTGTGAGGCAGTTG-3′, reverse: 5′-TGGTCACTCACATGGATGATCTG-3′. Primers for the human HK2 gene were forward: 5′-GAGCCACCACTCACCCTACT-3′, reverse: 5′-CCAGGCATTCGGCAATGTG-3′. Primers for the human β-actin gene were forward: 5′-GGCACCACACCTTCTACAATG-3′, reverse: 5′-TAGCACAGCCTGGATAGCAAC-3′. The relative mRNA levels of target genes were obtained by using the 2−ΔΔCt method with all assays performed in triplicate.
Cell Proliferation, Invasion, and Colony Formation Assay
Cell proliferation and colony formation assays were performed as previously described.20, 21 Invasion of MGC80-3 cells was detected by a transwell system. About 2 × 105 MGC80-3 cells were seeded on the top chamber (diluted with 100 µL of DMEM) of the Polycarbonate Membrane Transwell Inserts (Corning Incorporated, Corning, NY) precoated with diluted Matrigel Basement Membrane Matrix (BD Biosciences, USA) according to the Product Specification Sheet. A total of 600 µL of DMEM containing 10% FBS was added in the bottom chamber in advance. After incubation for 20 h, the nonpenetrating cells on the upper side of the filters were removed by “scrubbing” with cotton-tipped swabs. Penetrated cells on the lower side were fixed, stained, and scored in ten randomly chosen microscopic fields to measure the invasion of MGC80-3 cells.
Western Blot Assay
Western blot assay was performed as described previously.22 The specific primary antibodies used in these assays including antibodies against HK2 (11 097-1-AP, Proteintech, Chicago, USA), and β-actin (TA-09, ZSGB-BIO, Beijing, China).
Protein Extraction
MGC80-3 cells added in lysis buffer (8 m urea, 1% Protease Inhibitor Cocktail) were sonicated on ice three times using a high intensity ultrasonic processor (Scientz). After centrifugation at 12 000 × g for 10 min at 4 °C, the remaining cell debris was removed. In the end, the supernatant was transferred into a new Eppendorf tube and the protein concentration was determined using a BCA kit according to the manufacturer's instructions.
Trypsin Digestion
Dithiothreitol was added to the protein solution to a final concentration of 5 mm and reduced at 56 °C for 30 min. Iodoacetamide was then added to a final concentration of 11 mm and incubated for 15 min at room temperature in darkness. The protein sample was then added 100 mm TEAB until the urea concentration was diluted to <2 m. Finally, trypsin was added at a mass ratio of 1:50 trypsin-to-protein for the first digestion overnight at 37 °C and 1:100 trypsin-to-protein for a 4 h digestion secondly.
TMT Labeling
The trypsin-digested peptide was desalted with Strata X C18 (Phenomenex) and vacuum-dried. The peptide was solubilized with 0.5 m TEAB and labeled according to the TMT kit (Thermo) instructions.
HPLC Fractionation
The tryptic peptides were fractionated by high pH reverse phase HPLC and the used column was Agilent 300Extend C18 (5 µm particles, 4.6 mm ID, 250 mm length). First, peptides were separated into 60 fractions with a gradient of 8–32% acetonitrile, and the pH was 9.0, for 60 min. Then, the peptides were combined into 18 fractions and dried by vacuum centrifuging.
LC–MS/MS Analysis
The tryptic peptides were dissolved in 0.1% formic acid (solvent A) and separated using an EASY-nLC 1000 UPLC system. The gradient consisted of an increase from 6% to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, 23% to 35% in 8 min and climbing to 80% in 3 min, and then holding at 80% for the last 3 min, all at a constant flow rate of 400 nL min–1.
The peptides separated by an UPLC system were injected to nanospray ionization source for ionization and then analyzed by MS/MS in Q Exactive Plus (Thermo). The electrospray voltage applied was 2.0 kV. The primary MS scan range was 350 to 1800 m/z, and intact peptides were detected at a resolution of 70 000 in the Orbitrap. Peptides for MS/MS were then selected using normalized collisional energy setting as 28 and the ion fragments were detected at a resolution of 17 500 in the Orbitrap. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control was set at 5E4. The fixed first mass was set as 100 m/z.
Database Search
The resulting MS/MS data were analyzed through Maxquant search engine (v.1.5.2.8), and tandem mass spectra were searched against Homo sapiens database concatenated with reverse decoy database. Trypsin/P was specified as a cleavage enzyme allowing up to two missing cleavages. The mass tolerance for precursor ions in first and main search was set as 20 and 5 ppm, respectively. In addition, the mass tolerance was set as 0.02 Da for fragment ions. Carbamidomethyl on Cys was specified as fixed modification, while oxidation on Met and acetylation on protein N-terminal were specified as variable modifications. FDR was adjusted to <1% and minimum score for peptides was set >40.
Bioinformatic Analysis of Differentially Expressed Proteins
GO annotation proteome was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA). Then proteins were classified by GO annotation (http://www.geneontology.org/). The wolfpsort (https://wolfpsort.hgc.jp/) database was used to predict subcellular localization. Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) database was used to annotate protein pathways. The InterPro database was used for protein domain functional description for identified proteins.
Statistical Analysis
Data were presented as mean ± SEM. Quantitative variables between two groups were evaluated with two-tailed Student's t-test. Data were statistically analyzed by GraphPad Prism 5 software (GraphPad, CA, USA). A p-value < 0.05 was considered statistically significant, and p-value used in all analyses was two-tailed.
Data Availability
The mass spectrometry proteomics data have been deposited in ProteomeXchange Consortium (http://www.proteomexchange.org) via PRIDE partner repository with the dataset identifier PXD015813. The data can be accessed using the following details: Username: [email protected]; password: 0wkkJ6bD.
Acknowledgements
The present study was supported by the Natural Science Foundation of Shandong Province (grant no. ZR2017MH035), the Medical Science and Technology Development Plan of Shandong Province (grant no. 2016WS0413), and the National Natural Science Foundation of China (no. 81772626).
Conflict of Interest
The authors declare no conflict of interest.