DNA Methylation Adds Prognostic Value to Minimal Residual Disease Status in Pediatric T-Cell Acute Lymphoblastic Leukemia
Grant sponsor: Swedish Cancer Society; Grant sponsor: Swedish Research Council; Grant sponsor: Swedish Childhood Cancer Foundation; Grant sponsor: Medical Faculty of Umeå University; Grant sponsor: Danish Childhood Cancer Foundation; Grant sponsor: Lion's Cancer Research Foundation, Umeå; Grant sponsor: Umeå Paediatric Clinic Research Foundation; Grant sponsor: Magnus Bergvalls stiftelse; Grant sponsor: Uppsala-Umeå Comprehensive Cancer Consortium.
Conflict of interest: Nothing to declare.
[This article was corrected on 23 June 2016 after initial online publication because the copyright line needed to be updated.]
Abstract
Background
Despite increased knowledge about genetic aberrations in pediatric T-cell acute lymphoblastic leukemia (T-ALL), no clinically feasible treatment-stratifying marker exists at diagnosis. Instead patients are enrolled in intensive induction therapies with substantial side effects. In modern protocols, therapy response is monitored by minimal residual disease (MRD) analysis and used for postinduction risk group stratification. DNA methylation profiling is a candidate for subtype discrimination at diagnosis and we investigated its role as a prognostic marker in pediatric T-ALL.
Procedure
Sixty-five diagnostic T-ALL samples from Nordic pediatric patients treated according to the Nordic Society of Pediatric Hematology and Oncology ALL 2008 (NOPHO ALL 2008) protocol were analyzed by HumMeth450K genome wide DNA methylation arrays. Methylation status was analyzed in relation to clinical data and early T-cell precursor (ETP) phenotype.
Results
Two distinct CpG island methylator phenotype (CIMP) groups were identified. Patients with a CIMP-negative profile had an inferior response to treatment compared to CIMP-positive patients (3-year cumulative incidence of relapse (CIR3y) rate: 29% vs. 6%, P = 0.01). Most importantly, CIMP classification at diagnosis allowed subgrouping of high-risk T-ALL patients (MRD ≥0.1% at day 29) into two groups with significant differences in outcome (CIR3y rates: CIMP negative 50% vs. CIMP positive 12%; P = 0.02). These groups did not differ regarding ETP phenotype, but the CIMP-negative group was younger (P = 0.02) and had higher white blood cell count at diagnosis (P = 0.004) compared with the CIMP-positive group.
Conclusions
CIMP classification at diagnosis in combination with MRD during induction therapy is a strong candidate for further risk classification and could confer important information in treatment decision making.
Abbreviations
-
- CIMP
-
- CpG island methylator phenotype
-
- CIR
-
- cumulative incidence of relapse
-
- CIR3y
-
- 3-year cumulative incidence of relapse
-
- CpG
-
- cytosine-phosphate-guanine dinucleotide
-
- EGIL
-
- European group for the immunological classification of leukemias
-
- ETP
-
- early T-cell precursor
-
- HRM
-
- high-resolution melting curve
-
- MRD
-
- minimal residual disease
-
- OS
-
- overall survival
-
- pOS3yr
-
- 3-year overall survival rate
-
- WBC
-
- white blood cell
INTRODUCTION
T-cell acute lymphoblastic leukemia (T-ALL) accounts for 10–15% of childhood ALL, but is more common among adolescents and adults.1 The malignant transformation involves several genomic changes altering the normal control of T-cell development and proliferation.2 Although the pathogenesis of T-ALL has been extensively studied, few clinically useful prognostic markers exist beyond minimal residual disease (MRD) status during the first months of therapy.3
During the last decade, “epigenetics” (that is, functionally relevant changes in the genome that might influence gene expression without affecting the nucleotide sequence), has emerged as an important player in tumor development. One general finding during malignant transformation is a decrease in global DNA methylation, contributing to genomic instability, and an increase in promoter associated CpG island methylation associated with downregulation of tumor suppressor genes.4 We and others have demonstrated both subtype classification and prognostic relevance of aberrant DNA methylation patterns in various hematological disorders.5-9 However, the definition of methylation classification into CpG island methylator phenotype (CIMP) subgroups is poorly defined and ranges from classification of single predefined genes to unsupervised genome-wide methylation profiling in different studies.10 A few other papers have been published on pediatric T-ALL, methylation, and prognosis, but these studies have classified CIMP status based on single selected genes and are not comparable with our genome-wide approach.11, 12
Based on our previous finding of a strong prognostic significance of promoter-associated DNA methylation in Swedish T-ALL patients treated according to the Nordic Society of Pediatric Hematology and Oncology (NOPHO) ALL 1992/2000 protocols,6 we here explore its prognostic relevance in a new independent Nordic cohort of patients treated with the currently used NOPHO ALL 2008 protocol. In contrast to the ALL 1992/2000 protocols, the ALL 2008 protocol uses postinduction MRD levels for risk group stratification.1
METHODS
T-Cell Acute Leukemia and Control Samples
Between July 2008 and March 2013, 113 children (age <18 years) were diagnosed in the Nordic countries with T-ALL and treated according to the common NOPHO ALL 2008 protocol.1 Sixty-five diagnostic bone marrow/peripheral blood samples were available in the NOPHO leukemia biobank in Uppsala, Sweden, and were analyzed for methylation status. The T-ALL diagnosis was in each case based on morphology and flow cytometric immunophenotyping. Cytogenetic aberrations were explored by G-band karyotyping and targeted FISH analysis.1 Clinical data including white blood cell (WBC) count, immunophenotype, cytogenetic aberrations at diagnosis, and MRD status at treatment day 29 (end of induction) were evaluated in relation to methylation status at diagnosis. MRD was monitored by PCR and/or flow cytometry. PCR analysis of clonal gene rearrangements was recommended for MRD quantification in T-cell ALL and such MRD data 13 were used when available (n = 41). However, if no PCR-based MRD was available, flow cytometric quantification of MRD 14 was used (n = 20). Four cases lacked both PCR and flow MRD data and were excluded from the survival analyses that included MRD.
T-cell maturation stage was evaluated in the diagnostic samples. The European Group for the Immunological Classification of Leukemias (EGIL) criteria were used as defined by Bene et al.3 The immature subgroup of T-ALL described by Coustan-Smith et al. 15 as “early T-cell precursor” (ETP) is characterized by hematopoietic stem cell (HSC) and myeloid progenitor markers.3, 15-17
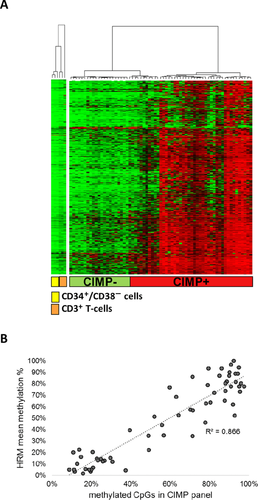
Publicly available methylation data (NCBI GEO database, GSE49618) of sorted T-cells (CD3+) and HSCs (CD34+, CD38–) from healthy donors were used as reference samples in the methylation heat map (Fig. 1A) to illustrate methylation profiles of normal immature and mature hematopoietic cells.
The study was approved by the regional and/or national ethics committees, and the patients and/or their guardians provided informed consent in accordance with the Declaration of Helsinki.
Genome Wide CpG Site Methylation Profiling
DNA was extracted by the AllPrep DNA/RNA kit (Qiagen, Hilden, Germany) and diagnostic T-ALL samples were analyzed by the HumMeth450K methylation array (lllumina, San Diego, CA) covering 485,577 CpG sites. Bisulfite conversion was performed according to manufacturer's manual (Zymo Research, Irvine, CA) and array analysis including preprocessing and normalization was performed as previously described.18 CpG probes that align to multiple loci in the genome or were located less than 3 bp from a known single nucleotide polymorphism were excluded.19 The fluorescence intensities were extracted using the Methylation Module (1.9.0) in the Genome Studio software (V2011.1). The methylation level (β value) of each CpG site ranged from 0 (no methylation) to 1 (complete methylation). The quality of each individual array was evaluated with built-in controls. Two replicate samples were included to assess interassay reproducibility (R2 = 0.97–0.99). The methylation array data were deposited in the NCBI GEO database, GSE69954. The normalized β values were used as measures of methylation levels and downstream analysis of the data was performed using R (v2.15.0).
All samples were CpG island methylator phenotype (CIMP) classified according to the previously described CIMP panel 6 using 1,293 CpG sites present in the HumMeth450K array. In short, in order to identify T-ALL methylation subgroups, the CIMP panel was defined as the most variable CpG sites in the 27K array (Illumina) within diagnostic T-ALL samples. The CpG sites within the CIMP panel were characterized by being located within CpG islands and associated with polycomb-target genes.6
In order to standardize CIMP classification independent of clustering, a cut-off level for “percentage of methylated CpGs within CIMP panel” was defined for the CIMP subgroups. Diagnostic T-ALL samples with >40% methylated CpG sites (each CpG site was considered methylated if the beta value was >0.4) in the CIMP panel (1293 CpGs) were classified as CIMP positive, whereas samples with ≤40% methylated sites were denoted CIMP negative. The cut-off was set to reflect the previously identified clusters to be the most discriminating with respect to prognosis.6
Verification of Methylation Array Data by High-Resolution Melting Curve Analysis
High-resolution melting (HRM) assays were designed for a selection of genes in 1,293 CpG site CIMP panel, representing CpG sites with distinct differences in methylation levels between CIMP subgroups in the array. The genes included KLF4, TFAP2C, IGFBP3, WNT3A, GATA4, and EYA4. Each 25 μl HRM reaction mix contained 1X Epitect HRM PCR Mastermix (Qiagen), 0.75 μM of forward and reverse primers (Supplementary Table SI), and 10 ng DNA template. The analysis was run in a RotorGene instrument (Qiagen) as follows: 95°C for 5 min, followed by 35 cycles of 95°C for 10 sec, 55°C for 30 sec, and 72°C for 20 sec. HRM was conducted by melting from 60 to 90°C, rising by 0.1° each step. A standard curve was prepared by mixing 100% methylated DNA (M.SssI treated) in different ratios with DNA from mitogen (wheat germ agglutinin) stimulated primary lymphoblast T-cell cultures P7/R2 (theoretically 0% methylated mononuclear cells). The methylation level of each gene region covered in the HRM assay was estimated in relation to the standard curve, and a mean methylation level (%) of the six-gene HRM panel was calculated. DNA was available from 63 of 65 array-analyzed T-ALL samples. Data were analyzed using Rotor-Gene® software v1.7 (Qiagen).
Statistical Analysis
The Statistical Package for the Social Sciences (SPSS Inc., Chicago, IL) software for Macintosh 22 was used for the statistical analyses. The chi-square/Fisher's exact test was used to compare differences between subgroups among categorical variables and the Mann–Whitney U test was used for continuous variables. Estimates of 3-year cumulative incidence of relapse (pCIR3yr) and overall survival (pOS3yr) rates were calculated using the Kaplan–Meier method and the subgroups listed in tables were compared using the log rank test. The significance level used in all tests was 0.05. Time in first remission (CR1) was defined as time (month) from diagnosis until first event. In CIR analysis, relapse was the endpoint. In the overall survival (OS) analysis, death from any cause was the endpoint. Moreover, the cumulative incidence of death in remission (CIDCR1) was compared between CIMP subgroups. The NOPHO leukemia registry is updated annually, and follow-up data were extracted from the registry as of May 2015.
RESULTS
Demographic Data
One hundred thirteen children (age ≤18 years) were diagnosed with T-ALL in the Nordic countries between July 2008 and March 2013, and treated according to the Nordic study protocol NOPHO ALL 2008. Clinical response to induction therapy with dexamethasone, vincristine, doxorubicin, and intrathecal methotrexate 1 was evaluated by MRD at day 29. The response to induction therapy determined whether the patients were assigned to antimetabolite-based intermediate risk therapy (MRD < 0.1%) or intensive myelosuppressive high risk (MRD ≥0.1%) block therapy, respectively.1, 20
A total of 65 diagnostic T-ALL samples in the NOPHO biobank were available for methylation analysis. Apart from higher WBC counts at diagnosis for those included (P = 0.03), there were no significant differences between the analyzed (n = 65) and not analyzed (n = 48) samples; that is, no statistically significant differences were found regarding age, gender, or MRD day 29 status (fraction <0.1%/≥0.1%; Table I). CIR and OS analysis confirmed that the 65 patients with available samples in the NOPHO biobank were representative for all pediatric T-ALL cases diagnosed in the Nordic countries during the study period with no difference in pCIR3yr between patients analyzed for methylation (n = 65) or not analyzed (n = 48, 15% vs. 13%, P = 0.82) or pOS3yr (79% vs. 79%, P = 0.85; Table I).
| CIMP Analyzed | Not analyzed | Total | ||
|---|---|---|---|---|
| N = 65 | N = 48 | P-value | N = 113 | |
| Gender male/female | 43/22 | 35/13 | ns | 78/35 |
| Median age (range, years) | 7 (1–17) | 7 (2–17) | ns | 7 (1–17) |
| Median WBC x 109/l (range) | 150 (1.6–983) | 67.1 (0.7–938) | 0.03 | 98.3 (0.7–983) |
| MRD day 29 <0.1%/≥0.1%/NA | 28/33/4 | 29/18/1 | ns | 57/51/5 |
| pCIR3y (SE) | 0.15 ± 0.05 | 0.13 ± 0.06 | ns | 0.15 ± 0.04 |
| pOS3y (SE) | 0.79 ± 0.05 | 0.79 ± 0.07 | ns | 0.79 ± 0.04 |
- NA, not analyzed; ns, not significant; pCIR3y, 3-year cumulative incidence of relapse; pOS3y, 3-year overall survival rate; SE, standard error.
CIMP Classification and Verification
The diagnostic T-ALL samples were analyzed by HumMeth450K arrays, and CpG island methylation phenotype (CIMP) classified by a previously defined panel of 1,293 CpG sites.6 Twenty-five T-ALL patients were classified as CIMP negative and 40 were classified as CIMP positive (Fig. 1A and Supplementary Table SII). The CIMP-negative samples had a methylation profile very similar to normal CD3+ T-cells and CD34+ HSCs (Fig. 1A and Supplementary Table SII).
The validity of the methylation arrays was determined by HRM analysis of six gene regions covering a selection of CpG sites with distinct differences in methylation levels between CIMP subgroups in the array CIMP panel. The mean HRM methylation level (%) of the six-gene HRM-CIMP panel correlated well with the percentage of methylated CpG sites in the array-CIMP panel (R2 = 0.87, Fig. 1B).
CIMP Classes and Clinical Characteristics Including T-Cell Maturation Stage
CIMP-negative (n = 25) and CIMP-positive (n = 40) T-ALL patients showed no significant differences regarding gender, age, or MRD status (<0.1%/≥0.1%) at day 29, but higher WBC values were found in CIMP-negative compared to CIMP-positive cases (median WBC: 228 vs. 77 × 109/l; P = 0.03) (Table II).
| CIMP+ | CIMP– | Total | |||
|---|---|---|---|---|---|
| N = 40 | N = 25 | P-value | N = 65 | ||
| Gender | male/female | 27/13 | 16/9 | ns | 43/22 |
| Median age (range, years) | 7 (2–17) | 8 (1–15) | ns | 7 (1–17) | |
| Median WBC x 109/l (range) | 77.1 (1.6–825) | 228 (6.9–983) | 0.03 | 150 (1.6–983) | |
| MRD day 29 | <0.1%/≥0.1%/NA | 18/18/4 | 10/15/0 | ns | 28/33/4 |
| ETP phenotype1 | Yes/No/NA | 6/28/6 | 2/20/3 | ns | 8/48/9 |
| EGIL class2 | Immature/cortical /mature/NA | 13/19/2/6 | 7/11/4/3 | ns | 20/30/6/9 |
| Follow-up status | CR1 (median follow up; range, months) | 35 (50; 2–76) | 17 (58; 24–73) | 52 (52; 2–76) | |
| Relapse (median time to relapse, months) | 2 (18) | 7 (13) | 9 (13) | ||
| DCR1 | 2 | 1 | 3 | ||
| Induction failure | 1 | 0 | 1 | ||
| Resistant disease | 0 | 0 | 0 | ||
| Dead/alive | 5/35 | 8/17 | 13/52 | ||
| pCIR3y (SE) | 0.06 ± 0.04 | 0.29 ± 0.09 | 0.01 | 0.15 ± 0.05 | |
| pCIDCR13y (SE) | 0.05 ± 0.04 | 0.05 ± 0.05 | ns | 0.05 ± 0.03 | |
| pOS3y (SE) | 0.87 ± 0.06 | 0.68 ± 0.1 | 0.08 | 0.79 ± 0.05 |
Among the 65 methylation analyzed patients, 56 had sufficient data to allow for T-cell maturation stage classification (EGIL class and ETP phenotype). We found no significant association between T-cell maturation stage (EGIL class or ETP phenotype) and CIMP class (Table II).
Survival Analysis
The pCIR3yr analysis of patients classified according to CIMP status showed significant differences (P = 0.01) with the best prognosis for CIMP-positive cases (pCIR3yr 6%) compared to CIMP-negative cases (pCIR3yr 29%) (Fig. 2A and Table II). The pOS3yr also differed between the CIMP groups (pOS3yr: 87% for CIMP positive and 68% for CIMP negative), but was not significant (P = 0.08) (Fig. 2A and Table II). The cumulative incidence of death in remission (pCIDCR13yr) did not differ between CIMP subgroups (P = 0.86) (Table II) and was therefore not corrected for.

MRD at day 29 is used for risk group stratification in the current NOPHO ALL 2008 protocol. The pCIR3yr and pOS3yr analysis of the 61 samples with available MRD data showed that MRD ≥0.1% day 29 identified individuals with poor outcome (pCIR3yr MRD ≥0.1%, 30% and MRD <0.1%, 0%, P = 0.003, and pOS3yr MRD ≥0.1%, 65% and MRD <0.1%, 100%, P = 0.001; Fig. 2B).
By combining CIMP status at diagnosis and MRD level at day 29, this prognostic information was enhanced (P < 0.001; Fig. 2C). The MRD ≥0.1%/CIMP negative group could be identified as a group with very poor prognosis (pCIR3yr 50% and pOS3yr 45%), whereas the MRD ≥0.1%/CIMP-positive patients had a much better outcome (pCIR3yr 12% and pOS3yr 83%) (P = 0.02 [CIR], P = 0.03 [OS]; Table III). Interestingly, CIMP status did not play a role for patients with low MRD, since there were no events in the MRD <0.1% group, irrespective of the CIMP status (pCIR3yr 100% and pOS3yr 100%) (Fig. 2C and Table III).
| MRD ≥ 0.1% | MRD < 0.1% | ||||||
|---|---|---|---|---|---|---|---|
| CIMP+ (N = 18) | CIMP– (N = 15) | P-value | CIMP+ (N = 18) | CIMP– (N = 10) | P-value | ||
| Gender | male/female | 13/5 | 10/5 | ns | 12/6 | 6/4 | ns |
| Median age (range, years) | 11.5 (2–17) | 7 (1–15) | 0.02 | 5 (2–14) | 12 (1–15) | ns | |
| Median WBC x 109/l (range) | 72 (3–560) | 241 (52–983) | 0.004 | 74 (4–825) | 126 (7–492) | ns | |
| ETP1 | Yes/no/NA | 3/14/1 | 1/13/1 | ns | 2/11/5 | 1/7/2 | ns |
| EGIL2 | Immature/cortical /mature/NA | 6/9/2/1 | 4/8/2/1 | ns | 5/8/0/5 | 3/3/2/2 | ns |
| Therapy stratification after day 29 | Standard risk | 0 | 0 | 0 | 0 | ||
| Intermediate risk | 0 | 0 | 17 | 9 | |||
| High-risk chemo | 16 | 13 | 1 | 1 | |||
| High-risk SCTCR1 | 2 | 2 | 0 | 0 | |||
| Follow-up status | CR1 | 15 | 7 | 18 | 10 | ||
| Induction failure | 0 | 0 | 0 | 0 | |||
| Resistant disease | 0 | 0 | 0 | 0 | |||
| Relapse | 2 | 7 | 0 | 0 | |||
| DCR1 | 1 | 1 | 0 | 0 | |||
| Dead/alive | 3/15 | 8/7 | 0/18 | 0/10 | |||
| pCIR3y (SE) | 0.12 ± 0.08 | 0.50 ± 0.14 | 0.02 | 0 | 0 | ns | |
| pOS3y (SE) | 0.83 ± 0.09 | 0.45 ± 0.13 | 0.03 | 1 | 1 | ns | |
When comparing the MRD ≥0.1%/CIMP negative group with the MRD ≥0.1%/CIMP positive group for clinical data including age, gender, WBC at diagnosis, ETP, and EGIL status, only age and WBC differed significantly. Thus, the MRD ≥0.1%/CIMP negative group had higher WBC count (Median: 241 × 109/l vs. 72 × 109/l, P = 0.004) and were younger (Median: 7.0 vs. 11.5 years, P = 0.02) at diagnosis compared with the MRD ≥0.1%/CIMP positive group (Table III).
To further study their relation, WBC were plotted together with CIMP and MRD status (Fig. 3) showing that our identified poor prognostic, MRD ≥0.1%/CIMP negative group, was characterized by high WBC counts at diagnosis.

DISCUSSION
Even though the outcome of pediatric T-ALL has improved significantly over the last decades, the high intensive chemotherapy treatment currently used may cause short- or long-term adverse effects. Furthermore, the cure rate of relapsed T-ALL has remained dismal, calling for improved risk stratification that allow those with the highest risk of relapse to be allocated to novel treatment strategies and/or HSC transplantation in first remission.
We have previously shown a strong prognostic impact of DNA methylation classification into CIMP groups in pediatric T-ALL patients treated according to the NOPHO ALL 1992/2000 protocols.6 However, in those protocols the cure rate for T-cell ALL was generally poor. The present study in an independent Nordic cohort confirms that CIMP classification is a relevant clinical prognostic factor for childhood T-ALL, also in the setting of contemporary treatment programs that integrate MRD measurements in the risk group allocation. Although the overall pCIR rate has been improved with the current ALL2008 protocol, CIMP status at diagnosis was a significant prognostic factor. Of special importance is that a combination of CIMP status at diagnosis and MRD status at treatment day 29 could further identify patients with significantly different clinical outcomes. In the current NOPHO ALL 2008 protocol (NCT00816049) for T-ALL, MRD levels >0.1% at day 29 stratifies patients to an intensified treatment schedule.1 Whether a future combination of CIMP classification and postinduction MRD levels will allow down grading of CIMP-positive T-ALL patients to less intensive chemotherapy must be evaluated in prospective trials. Importantly, the prognosis for CIMP negative/MRD ≥0.1% patients is so dismal that novel treatment approaches are needed. Methylation status does not seem to influence the initial response to therapy since there were no significant difference in remaining leukemic cells after induction therapy (MRD status day 29) in CIMP subgroups. The vast majority of leukemic cells were eliminated by therapy regardless of methylation status. However, the relapse frequency was higher in the CIMP-negative leukemias. The reason for this we can only speculate upon but it might reflect different efficiency of eliminating leukemic initiating cells in CIMP-positive and CIMP-negative leukemias.
Our CIMP profile reflecting 1,293 CpGs is based on methylation array analysis, and is a robust and informative, but yet relatively expensive (≈ 350 USD/sample) and time consuming technique (3 days). As an alternative technique for CIMP classification and to confirm the array CIMP classification, we used HRM analysis based on six gene regions selected from the CIMP panel to represent CpG sites with distinct differences in methylation levels between the CIMP subgroups. The HRM classification correlated well with the array-based CIMP classification, and it can be run on single samples and be completed within 1 day. Therefore, HRM CIMP classification is an attractive alternative to array CIMP classification. However, the array analysis has its advantage in gaining information about genome-wide methylation patterns and can be used for additional analysis, including copy number alterations.21
Prognostic impact of DNA methylation signatures has also been observed in other hematological malignancies, including myelodysplasia, acute myeloid leukemia (AML), and BCP-ALL, and methylation modifiers play a therapeutic role.8, 22, 23 The prognostic relevance of CIMP classification in pediatric T-ALL shown in this paper confirms our previous finding in a separate T-ALL cohort. However, there are conflicting results of CIMP status and prognosis in pediatric T-ALL,11, 12 but these studies used 14–20 predefined genes for methylation classification in contrast to our genome-wide array approach. In order to gain the full potential of DNA methylation classification as a prognostic marker in T-ALL, further validation of larger cohorts is needed.
A deeper functional understanding of the complex role of DNA methylation aberrations in the development and progression of hematological malignancies is still missing.
Genetic subtypes of childhood ALL and AML have been associated with DNA methylation signatures.24-26 A recent study by Amabile et al. showed that in a murine chronic myeloid leukemia (CML) model, induction of BCR-ABL could trigger DNA methylation changes. Furthermore, it was shown that aberrant DNA methylation had the potential to contribute to leukemia progression in primary CML cells.27
Genome-wide methylation studies of the mouse hematopoietic system identified specific signatures associated with lineage commitment and T-cell maturation stage and showed that cells committed to a myeloid lineage had lower global methylation levels than cells committed to a lymphoid lineage.28 Immature T-ALL and ETP-ALL, with myeloid molecular characteristics, have been associated with poor outcome.15, 29-32 However, we found no significant association of CIMP subgroup with T-cell maturation stage based on ETP phenotype or EGIL classification.
The MRD ≥0.1%/CIMP negative group was associated with high WBC counts at diagnosis. WBC count is not used as a stratifier for therapy in T-ALL in the current protocol but analysis are ongoing whether or not it should be included in future risk assessments.33 The CIMP, MRD, and WBC factors partly covaried and it seems that combining two of these three factors may be useful to further define different risk groups. However, larger sample size is needed to study their relation/independence in detail by multivariate Cox regression analysis.
DNA methylation alterations have been shown to accumulate in cells over time,18, 34, 35 and our recent publication on a T-cell culture model for immortalization showed overlapping methylation alterations in long-term cultured immortalized cell cultures and CIMP-positive T-ALL.18 Age-related alterations in the DNA methylation patterns have been studied in the ontogeny of HSCs showing a significant global DNA hypermethylation in older HSCs. The same study found that hypermethylation occurred at polycomb repressive complex 2 (PRC2) target loci upon forced proliferation of HSCs.36 Our CIMP profile is enriched for CpG sites located in polycomb target genes.6 The fact that the CIMP-negative subgroup showed a similar methylation profile to normal CD3+ and CD34+/CD38– cells suggests that these cases might have undergone fewer rounds of replications.
Recurring loss-of-function mutations and deletions have been identified in the PRC2 components (EZH2, SUZ12, and EED) in pediatric T-ALL.32 In adult T-ALL, mutations in RUNX1 and the DNA methyltransferase DNMT3A gene were associated with poor prognosis.37, 38 Recently, DNMT3A mutated preleukemic HSCs have been identified in AML that were resistant to chemotherapy and persisted in remission, indicating that they might represent a reservoir from which relapse arises.39 It remains to be determined, if mutations or genetic aberrations in DNA methylation associated genes or oncogenes can be associated with CIMP subgroups.
To further gain insight into characteristics of CIMP-positive and CIMP-negative cells, the next step will be to perform transcriptome and genome sequence analysis of diagnostic and relapse samples and relate to DNA methylation and histone modification alterations. In vitro cytotoxicity analysis of CIMP-positive and CIMP-negative cells will gain deepened knowledge of drug resistance and cell signaling response in cells of different methylation status.
To conclude, in this collaboration study between the Nordic countries we show that DNA methylation patterns in diagnostic T-ALL samples hold important prognostic information. Of special interest was that CIMP status at diagnosis could separate postinduction MRD-positive (>0.1%, day 29) patients into two risk groups. Further dissection of the biology behind CIMP status will hopefully result in the identification of novel therapeutic targets.
ACKNOWLEDGMENTS
This study was supported by grants from the Swedish Cancer Society, the Swedish Research Council (Dnr 340-2013-5185), the Swedish Childhood Cancer Foundation, the Medical Faculty of Umeå University, the Danish Childhood Cancer Foundation, Lion's Cancer Research Foundation (Umeå), Umeå Paediatric Clinic Research Foundation, Magnus Bergvalls stiftelse, and Uppsala-Umeå Comprehensive Cancer Consortium. Financial support was provided through regional agreement between Umeå University and Västerbotten County Council on cooperation in the field of medicine, odontology, and health. We thank Helene Sandström for laboratory assistance.
Authors' contributions
M. B., Z. H., M. Hu., G. R., E. F., and S. D. conceived and designed the experiments; U. N. N., K. S., A. Å., J. K., H. O. M., H. M., M. He. were involved in collection of data; S. D. and Z. H. performed the experiments; M. B., Z. H., M. L., E. F., and S. D. analyzed the data; and M. B., Z. H., K. S., G. R., and S. D. wrote the paper with contribution from all coauthors.




