Proteomic and Metabolite Profiling Reveals Profound Structural and Metabolic Reorganization of Adipocyte Mitochondria in Obesity
Abstract
Objective
Previous studies have revealed decreased mitochondrial respiration in adipocytes of obese mice. This study aimed to identify the molecular underpinnings of altered mitochondrial metabolism in adipocytes.
Methods
Untargeted proteomics of mitochondria isolated from adipocytes and metabolite profiling of adipose tissues were conducted in diet-induced obese (DIO) and lean mice. Subcutaneous and intra-abdominal adipose tissues were studied to depict depot-specific alterations.
Results
In subcutaneous adipocytes of DIO mice, changes in proteins related to mitochondrial structure and function were observed. Mitochondrial proteins of the inner and outer membrane were strongly reduced, whereas proteins of key matrix metabolic pathways were increased in the obese versus lean state, as further substantiated by metabolite profiling. A pronounced decrease in the oxidative phosphorylation (OXPHOS) enzymatic equipment and cristae density of the inner membrane was identified. In intra-abdominal adipocytes, similar systematic downregulation of the OXPHOS machinery in obesity occurred, but there was no regulation of outer membrane or matrix proteins.
Conclusions
Protein components of the OXPHOS machinery are systematically downregulated in adipose tissues of DIO mice compared with lean mice. Loss of the mitochondrial OXPHOS capacity in adipocytes may aggravate the development of metabolic disease.
Study Importance
What is already known?
- ► Mitochondrial respiration and oxidative phosphorylation (OXPHOS) capacity of adipocytes are rapidly decreased after onset of high-fat diet feeding.
- ► In diet-induced obese (DIO) mice, mitochondrial content of adipocytes is reduced.
- ► Loss of OXPHOS functionality is aggravated during chronic DIO development.
What does this study add?
- ► In subcutaneous adipocytes of DIO mice, the abundance of mitochondrial membrane proteins is strongly downregulated, whereas matrix proteins are upregulated.
- ► Among the inner membrane proteins, components of the OXPHOS machinery are systematically downregulated in adipocyte mitochondria of DIO mice.
- ► Observed massive changes in molecular architecture are associated with corresponding changes in metabolite profiles of adipose tissues.
- ► Obesity causes an imbalance between functionally coupled pathways located in the inner membrane and the matrix.
Introduction
The impact of adipocyte mitochondria on cellular homeostasis, their relevance for systemic metabolic health, and the therapeutic potential to target adipocyte mitochondria for treatment are gaining attention (1). Several studies have addressed the relevance of mitochondria in adipose tissue (AT) for both cellular and systemic homeostasis (2-11). Importantly, changes in adipocyte mitochondria characteristics in obesity and/or metabolic diseases have been observed on the microscopic, the transcriptomic, the proteomic, and finally the functional level (5, 12-17). In our previous work, we observed a limitation in oxidative phosphorylation (OXPHOS) capacity in adipocytes in several murine models of obesity (11). Moreover, we identified differences in adipocyte mitochondrial OXPHOS capacity between subcutaneous and intra-abdominal fat that may be linked to the divergent metabolic risk associated with these types of body fat (18). Thus, in the obese state, alterations in adipocyte mitochondrial functions may aggravate the development of metabolic disease.
Here, we characterized the proteome of adipocyte mitochondria derived from both subcutaneous and intra-abdominal AT in an obesity background using diet-induced obese C57BL/6N mice as a model. We aimed to identify mitochondrial proteins and pathways differentially regulated in adipocytes in obese versus lean mice. For this purpose, mitochondria isolated from mature adipocytes of subcutaneous and intra-abdominal AT depots were subjected to an untargeted proteomics approach to identify diet-induced and depot-specific alterations. Overall, we report changes in mitochondrial architecture leading to a massive obesity-induced decrease in the OXPHOS machinery.
Methods
Mice
Experiments were conducted on male C57BL/6N mice housed in groups (three to five mice) in a specific pathogen-free environment on a 12-hour light-dark cycle at 22°C with ad libitum access to food and water. At an age of 7 weeks, chow diet (type M-Z; Ssniff, Soest, Germany) was replaced by a purified research control diet (CD) (12 kJ% fat, SS745-E702; Ssniff). After acclimatization for 1 week, mice were matched by body weight into CD and high-fat diet (HFD) groups (48 kJ% fat, SS745-E712; Ssniff). After 6 months of dietary intervention, body weight and composition (nuclear magnetic resonance [NMR] minispec; Bruker, Billerica, Massachusetts) were assessed and AT depots dissected (ethical approval by the Government of Upper Bavaria: Az. 55.2.1.54-2532-148-13).
Proteome analysis
Tissue dissection and isolation of adipocytes
Mice were killed by CO2 exposure and exsanguination. Posterior subcutaneous and intra-abdominal epididymal fat depots were dissected, weighed, and snap-frozen in liquid nitrogen. Large blood vessels were removed. Mature adipocytes were separated from other cell types by collagenase digestion as described in detail previously (collagenase type A; Roche Applied Science, Penzberg, Germany; 1g/L in Hank’s Balanced Salt Solution [14025-092; Gibco, Carlsbad, California] containing 4% bovine serum albumin [BSA]) (18). Regarding the low yield of mitochondria per milliliter of adipocyte suspension at mitochondrial isolation, fat depots of 8 to 12 mice were pooled.
Isolation of mitochondria
Adipocyte mitochondria were isolated as previously described (18). Mature adipocytes were transferred into a 15-mL glass-Teflon homogenizer and disrupted by five rotating strokes. Separation of mitochondria was achieved by differential centrifugation (4°C, 10 minutes at 800g, 10 minutes at 10,000g). Pellets were resuspended in KHE-BSA buffer (120 mM KCL, 5 mM KH2PO4, 3 mM HEPES, 1 mM EGTA, 0.5% BSA-fatty acid free, pH = 7.2). After completion of functional measurements (18), mitochondria were stored at −80°C. For the present analysis, mitochondria were carefully thawed on ice. Three independent mitochondrial preparations of each feeding group were combined. Thus, for each feeding group, we had one representative sample containing mitochondria of 27 to 29 mice. Mitochondria were pelleted (10 minutes at 16,000g, 4°C) and resuspended in BSA-free KHE buffer.
Sample preparation
Mitochondria were lysed in 8 M urea, 50 mM Tris/HCl (pH 8.0), and 1x protease inhibitor cocktail (Sigma-Aldrich, Munich, Germany). A total of 150 µg from each pool was used for three technical replicates. Proteins were reduced using 10 µM dithiothreitol alkylated with 50 mM chloroacetamide and digested in solution with trypsin (1:50 w/w enzyme:substrate ratio; Promega Corp., Madison, Wisconsin). Peptide desalting and dimethyl labeling were performed on self-packed C18 StageTips columns as previously described (19). Eluted peptides were combined, and further separation into 16 fractions using hydrophilic strong anion exchange separation was performed as described (20).
Proteome analysis using liquid chromatography-tandem mass spectrometry
Nanoflow liquid chromatography (LC)–tandem mass spectrometry (MS/MS) was performed by coupling an Eksigent NanoLC Ultra 1D+ (Eksigent, Dublin, California) to an Orbitrap Elite (Thermo Fisher Scientific, Bremen, Germany). Peptides were delivered to a trap column (100 μm × 2 cm, packed in-house with Reprosil-Pur C18-AQ, 5 μm resin; Dr. Maisch, Ammerbuch, Germany) at a flow rate of 5 μL/min in 100% solvent A (0.1% formic acid in high-performance LC (HPLC)–grade water). After 10 minutes of loading and washing, peptides were transferred to an analytical column (75 μm × 40 cm, packed in-house with Reprosil-Pur C18-Gold, 3 μm resin; Dr. Maisch) and separated using a 100-minute gradient from 2% to 32% of solvent B (0.1% formic acid, 5% dimethyl sulfoxide [DMSO] in acetonitrile; solvent A: 0.1% formic acid, 5% DMSO in water) at a 300-nL/min flow rate. The Orbitrap Elite was operated in data-dependent mode, automatically switching between MS and MS2. Full-scan MS spectra were acquired in the Orbitrap at 30,000 (mass/charge [m/z] 400) resolution after accumulation to a target value of 1,000,000. MS/MS were generated for up to 15 peptide precursors in the Orbitrap for fragmentation by using higher energy collision-induced dissociation at normalized collision energy of 30% and a resolution of 15,000 with a target value of 20,000 charges after accumulation for a maximum of 100 milliseconds.
Data analysis
The raw MS data were processed using MaxQuant software (version 1.4.1.2; https://www.maxquant.org/) for peak detection and quantification (21). MS/MS spectra were searched against the Uniprot mouse database (v06.06.14) using the Andromeda search engine (22) with the following search parameters: full tryptic specificity, up to two missed cleavage sites, carbamidomethylation of cysteine residues set as fixed modification, N-terminal protein acetylation and methionine oxidation as variable modifications, and dimethylation and dimethylation:2 H(4) of lysine residues and protein N-termini as labels for quantification. Mass spectra were recalibrated within MaxQuant (first search 20 parts per million [ppm] precursor tolerance) and subsequently researched with a mass tolerance of 6 ppm; fragment ion mass tolerance was set to 20 ppm. Search results were filtered to a maximum false discovery rate of 0.01 for proteins and peptides, and a peptide length of at least six amino acids was required.
Identification of mitochondrial proteins was achieved using the MitoCarta Inventory of Mammalian Mitochondrial Genes (Mouse MitoCarta2.0; Broad Institute, Cambridge, Massachusetts). Mitochondrial proteins identified by MitoCarta were sorted according to submitochondrial localization as follows: inner membrane, outer membrane, and matrix referring to entries in GeneCards (Gene Ontology, Cellular Components; LifeMap Sciences, Alameda, California).
Metabolite profiling
Tissue collection
Posterior subcutaneous and intra-abdominal epididymal fat depots were stored at −80°C. Methanol extracts from total fat were used to avoid metabolite degradation during adipocyte separation from the stromal vascular fraction.
Sample preparation and metabolite profiling using LC-MS/MS
Quantification of acylcarnitines was performed using LC-MS/MS according to our previously described method (23). Briefly, tissue specimens (50 mg) were homogenized manually in 1 mL of 100% ice-cold methanol, followed by sonification for 5 minutes. After centrifugation (15 minutes at 13,000g), 200 µL of supernatant was mixed with 10 µL of labeled internal standard (ChromSystems, Munich, Germany). Samples were vacuum dried. Acylcarnitines were derivatized to their butyl esters, as described by Gucciardi et al. (24), by addition of 100 µL of n-butanol containing 5% vol/vol acetyl chloride and incubation at 60°C for 20 minutes at 800 rpm. After evaporation to dryness, samples were reconstituted in 100 µL of methanol and transferred to glass vials. MS analysis was done on a triple-quadrupole QTRAP 5500 MS/MS (AB Sciex, Framingham, Massachusetts) coupled to a 1200 series HPLC system (Agilent, Santa Clara, California) using the parameters described in our method. Acylcarnitine concentrations were corrected for the exact amount of weighted tissue.
Statistics
Statistical analysis was performed using Microsoft Excel (Redmond, Washington). For proteomics, differential expression was assessed based on the normalized H/L ratios (i.e., the normalized ratio of the mass spectrometric signal of heavy [H] and light [L] dimethyl labeled peptides) provided by MaxQuant analysis with moderated t test and Benjamini–Hochberg adjusted P values. For metabolomics, single peak intensities were normalized to the total profile intensity. Differential acylcarnitine concentrations were assessed based on the HFD/CD ratio with t test. Significance was accepted at P < 0.05.
Results
Diet-induced obesity reorganizes adipocyte mitochondria toward a lower membrane to matrix ratio in subcutaneous adipocytes
We identified mitochondrial proteins and pathways differentially regulated in adipocytes in obese versus lean mice. After 6 months of HFD feeding, C57BL/6N mice were massively obese with a higher body and fat mass but comparable lean mass (Supporting Information Table S1). Tissue weights of the posterior subcutaneous and the intra-abdominal fat depots were significantly elevated. From these fat pads, mature adipocytes were separated from cells of the stromal vascular fraction by collagenase digestion and mitochondria isolated from mature adipocytes by differential centrifugation. An untargeted comparative proteomic approach was performed to gain comprehensive insights into obesity-induced changes in mitochondrial organization and physiology (Supporting Information Figure S1A; raw data provided in Supporting Information Table S7). Proteome analysis revealed similar diet-induced changes in both fat pads, with the most prominent changes in the posterior subcutaneous depot.
In mitochondria of subcutaneous fat, we identified 898 unique proteins with a broad range of molecular weight ranging from 5.38 kDa (caveolin) to 3,906.40 kDa (titin). The 334 proteins clearly annotated as mitochondrial (identified by Mouse MitoCarta2.0 annotation) constituted approximately one-half of total protein mass (HFD: 46.0% ± 2.6%; CD: 54.2% ± 4.3%; Supporting Information Figure S1B). Of proteins with unambiguous submitochondrial localization (236 of 334, 70.7%), a majority represented 142 mitochondrial inner membrane (MIM) proteins (60.2%), followed by 72 matrix (30.5%) and 22 mitochondrial outer membrane (MOM) proteins (9.3%) (Figure 1A).
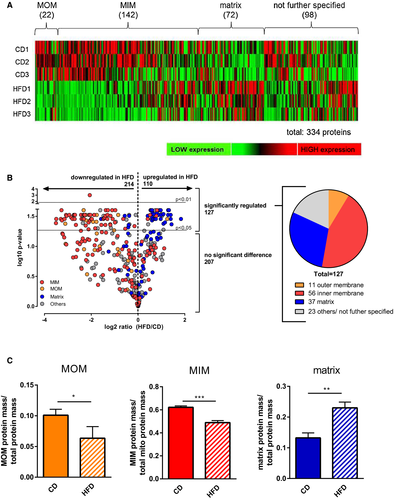
Comparing mitochondria from subcutaneous fat of obese versus lean mice, we observed a striking difference in the abundance of membrane versus matrix proteins (Figure 1). MIM and MOM proteins were strongly and consistently downregulated in obesity, whereas matrix proteins were increased. This was observed with respect to the abundance of individual proteins (Figure 1B) as well as regarding the overall contribution of MIM/MOM and matrix proteins to total protein mass (Figure 1C). Systematic downregulation was not restricted to mitochondrial proteins but was also observed for proteins with nonmitochondrial annotation according to MitoCarta. In total, 514 of 898 (57.2%) were downregulated, both mitochondrial and nonmitochondrial. The overall fold change of downregulation, however, was much higher for mitochondrial versus nonmitochondrial proteins (P < 0.001; Supporting Information Figure S1C). The altered ratio between membrane and matrix compartment proteins dominated all observed changes; of a total of 127 proteins significantly regulated (t test, Benjamini–Hochberg adjusted), 104 (81.9%) could be unambiguously assigned to MIM, MOM, or matrix (Figure 1B). This representation of differentially regulated MIM, MOM, and matrix proteins was similar in mitochondria of the intra-abdominal fat pad (Supporting Information Figure S2).
Taken together, diet-induced obesity caused a massive change in the balance between membrane and matrix protein pools and thus probably in overall mitochondrial architecture. To identify specific pathways affected by this reorganization, we continued by analyzing the MIM and MOM as well as the matrix in detail.
Overall reduction in MOM proteins in subcutaneous adipocytes
Our global, unbiased proteome analysis revealed a dramatic shift in the ratio between membrane and matrix protein abundance, demonstrating profound remodeling of the molecular architecture. Focusing on the MOM, this pattern remained evident. Of 11 significantly regulated MOM proteins, 10 were downregulated in HFD versus CD samples (Table 1). Most indicative for a reduced MOM expanse is the downregulation of all three isoforms of the voltage dependent anion channel (VDAC)/porin, as these beta-barrel pore proteins are commonly regarded as general markers of MOM expanse (25, 26).
| Gene name | Full name | HFD/CD ratio | Adj. P value |
|---|---|---|---|
| Samm50 | Sorting and assembly machinery component | −8.63 | 0.042 |
| Vdac3 | Voltage-dependent anion channel 3 | −5.01 | 0.025 |
| Vdac1 | Voltage-dependent anion channel 1 | −4.69 | 0.030 |
| Vdac2 | Voltage-dependent anion channel 2 | −4.65 | 0.037 |
| Dnajc11 | DnaJ heat shock protein family (Hsp40) member C11 | −4.18 | 0.001 |
| Cisd1 | CDGSH iron sulfur domain 1 | −3.79 | 0.039 |
| Cyb5 | Cytochrome b5 | −3.43 | 0.039 |
| Cyb5b | Cytochrome b5 type B | −3.40 | 0.030 |
| Cyb5r3 | Cytochrome b5 reductase 3 | −2.28 | 0.043 |
| Acsl1 | Acyl-CoA synthetase long-chain family member 1 | −1.80 | 0.030 |
| Pgam5 | Phosphoglycerate mutase family member 5 | 2.38 | 0.035 |
- Adj., adjusted; HFD, high-fat diet; CD, control diet.
The remaining downregulated proteins display divergent functions, such as activation of long-chain fatty acids (acyl-coenzyme A [CoA] synthetase long-chain family member 1), regulation of OXPHOS (CDGSH iron sulfur domain 1), or maintenance of mitochondrial cristae structure (sorting and assembly machinery component 50 [Samm50], DnaJ heat shock protein family [Hsp40] member C11 [Dnajc11]) and correct assembly of respiratory chain complexes (Dnajc11). Possibly, these proteins were identified because of their high abundance (such as VDAC) rather than their specific function.
In general, we observed an overall reduction of MOM proteins in adipocyte mitochondria from subcutaneous AT during obesity. Most striking is the systematic downregulation of VDAC, indicating a reduction in the expanse of the MOM.
Overall reduction in inner membrane proteins dominated by massive decrease in respiratory capacity in obesity
The strong shift in the proportion between membrane and matrix proteins in the obese state may be due to changes in mitochondrial morphology, a notion corroborated by reduced VDAC abundance in the MOM. Next, we focused on proteins located in the MIM of adipocyte mitochondria. Notably, 41 of 56 significantly regulated MIM proteins (73.2%) were identified to be part of the OXPHOS machinery (Figure 2). In mitochondria from the intra-abdominal fat pad, regulation was similar though less pronounced (Supporting Information Table S2). Strikingly, 40 of these respiratory chain subunits decreased in obesity, with reductions up to 10-fold. Similar to VDAC in the MOM, the majority of MIM proteins are part of OXPHOS complexes, and the overall reduction of these proteins points at a global decrease in MIM expanse. Therefore, obesity appears to both reduce MOM expanse and cristae density in adipocyte mitochondria. Both nuclear and mitochondrial DNA-encoded proteins contributed to reduced OXPHOS capacity in the obese state. We did not detect all subunits of OXPHOS protein complexes, but even if all of these would not be affected by the obese state, 54.5% of complex III subunits were still identified and downregulated, followed by complex I and complex II (both 50%), complex V (26%), and complex IV (25%).
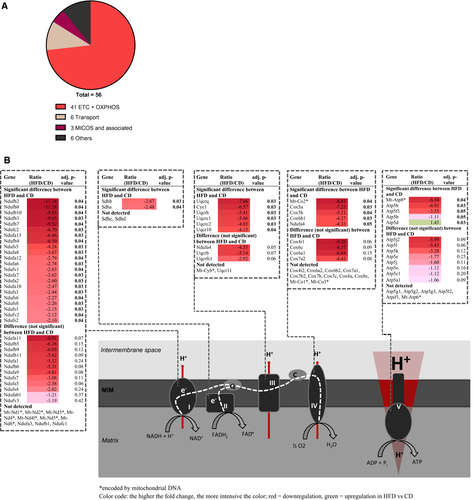
Beyond OXPHOS, 6 of 56 significantly regulated MIM proteins were identified as transmembrane carriers (Table 2), among them the adenine nucleotide translocase (Slc25a5). As substrate providers for mitochondrial ATP generation, their systematic downregulation (Table 2) further corroborates reduced OXPHOS capacity. Interestingly, three out of the total of seven subunits of the mitochondrial contact site and cristae organizing system (MICOS) were downregulated in response to HFD exposure (apolipoprotein O, apolipoprotein O-like, and the MIM protein; Table 2). The MICOS complex is involved in the formation and maintenance of cristae, cristae junctions, and contact sites to the MOM and is thus a key regulator of mitochondrial architecture (27). It interacts with VDAC, Samm50, and Dnajc11, all of which are MOM proteins downregulated in HFD versus CD mice (Table 1). Interaction of the latter two is required for cristae stability (28-31). Together, the specific downregulation of these structural maintenance proteins verifies the changes in mitochondrial architecture as indicated before by the global change in the membrane to matrix protein ratio.
| Gene name | Full name | HFD/CD ratio | Adj. P value |
|---|---|---|---|
| Transport across MIM | |||
| Mtch2 | Mitochondrial carrier 2 | −8.45 | 0.03 |
| Mpc2 | Mitochondrial pyruvate carrier 2 | −6.95 | 0.04 |
| Slc25a1 | Solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 | −5.33 | 0.00 |
| Slc25a10 | Solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 | −4.96 | 0.04 |
| Slc25a20 | Solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 | −4.32 | 0.04 |
| Slc25a5 | Solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 | −4.10 | 0.03 |
| MICOS | |||
| Apoo | Apolipoprotein O | −4.77 | 0.03 |
| Apool | Apolipoprotein O-like | −4.72 | 0.05 |
| Immt | Inner membrane mitochondrial protein | −1.96 | 0.03 |
| Others | |||
| Gpd2 | Glycerol phosphate dehydrogenase 2, mitochondrial | −8.28 | 0.03 |
| Abcd3 | ATP-binding cassette, sub-family D (ALD), member 3 | −3.93 | 0.04 |
| Chdh | Choline dehydrogenase | −2.96 | 0.04 |
| Aldh3a2 | Aldehyde dehydrogenase family 3, subfamily A2 | −1.98 | 0.04 |
| Aifm1 | Apoptosis-inducing factor, mitochondrion-associated 1 | −1.46 | 0.05 |
| Fech | Ferrochelatase | 1.33 | 0.04 |
- Adj., adjusted; CD, control diet; HFD, high-fat diet; MICOS, mitochondrial contact site and cristae organizing system; MIM, mitochondrial inner membrane.
In summary, we observed a significant reduction in MIM proteins in adipocytes of obese mice. First and foremost, we detected consequent downregulation of OXPHOS complexes but also MIM transporters, proteins of the MICOS complex, and some MOM partners, thus affirming altered mitochondrial architecture and reduced cristae density.
Obesity increases matrix protein abundance in adipocyte mitochondria
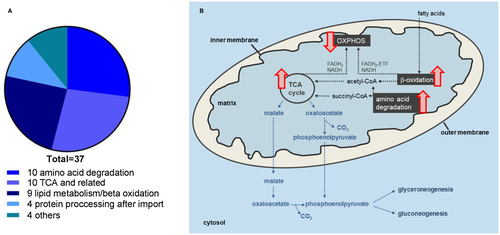
The abundance of both MIM and MOM proteins was strongly reduced in adipocyte mitochondria of diet-induced obese mice. Conversely, the amount of mitochondrial matrix proteins generally increased. The reduction in membrane proteins was contributed by the most abundant proteins (VDAC in MOM and OXPHOS complexes in MIM), indicating a major change in mitochondrial structure. Similarly, most of the identified matrix proteins (29 of 37) belonged to central bioenergetics pathways catabolizing the macronutrients carbohydrates, fat, and protein, including tricarboxylic acid (TCA) cycle, beta-oxidation, and amino acid degradation (Figure 3A and Table 3). Nearly all of these metabolic proteins were upregulated in response to HFD feeding (24 of 29). Again, this dominance of abundant pathways indicates structural alterations in adipocyte mitochondria caused by excessive energy storage.
| Gene name | Full name | HFD/CD ratio | Adj. P value |
|---|---|---|---|
| Amino acid degradation | |||
| Prodh | Proline dehydrogenase | −4.01 | 0.035 |
| Dbt | Dihydrolipoamide branched-chain transacylase E2 | −2.99 | 0.035 |
| Aldh4a1 | Aldehyde dehydrogenase 4 family member A1 | 1.33 | 0.031 |
| Mccc1 | Methylcrotonoyl-CoA carboxylase 1 (alpha) | 1.52 | 0.033 |
| Mut | Methylmalonyl-CoA mutase | 1.54 | 0.028 |
| Pccb | Propionyl-CoA carboxylase, beta polypeptide | 1.75 | 0.025 |
| Pcca | Propionyl-CoA carboxylase, alpha polypeptide | 1.96 | 0.035 |
| Oat | Ornithine aminotransferase | 2.13 | 0.042 |
| Auh | AU RNA binding protein/enoyl-CoA hydratase | 2.63 | 0.030 |
| Hibadh | 3-Hydroxyisobutyrate dehydrogenase | 2.63 | 0.035 |
| Fatty acid beta-oxidation /lipid catabolism | |||
| Ehhadh | Enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase | −1.48 | 0.031 |
| Etfb | Electron transferring flavoprotein, beta polypeptide | 1.69 | 0.035 |
| Acat1 | Acetyl-CoA acetyltransferase 1 | 1.72 | 0.031 |
| Acadl | Acyl-CoA dehydrogenase, long chain | 1.75 | 0.028 |
| Echs1 | Enoyl-CoA hydratase, short chain, 1, mitochondrial | 1.85 | 0.030 |
| Etfa | Electron transferring flavoprotein, alpha polypeptide | 1.89 | 0.025 |
| Hadh | Hydroxyacyl-CoA dehydrogenase | 2.13 | 0.029 |
| Acot9 | Acyl-CoA thioesterase 9 | 2.44 | 0.030 |
| Acadm | Acyl-CoA dehydrogenase, medium chain | 2.50 | 0.025 |
| TCA cycle and related | |||
| Dlst | Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | −1.33 | 0.048 |
| Pdha1 | Pyruvate dehydrogenase E1 alpha 1 | −1.32 | 0.044 |
| Idh2 | Isocitrate dehydrogenase 2 (NADP+), mitochondrial | 1.52 | 0.025 |
| Pdhx | Pyruvate dehydrogenase complex, component X | 1.52 | 0.037 |
| Mdh2 | Malate dehydrogenase 2, NAD, mitochondrial | 1.59 | 0.029 |
| Idh3a | Isocitrate dehydrogenase 3 (NAD+) alpha | 1.64 | 0.035 |
| Suclg1 | Succinate-CoA ligase, GDP-forming, alpha subunit | 1.75 | 0.025 |
| Cs | Citrate synthase | 1.96 | 0.025 |
| Glud1 | Glutamate dehydrogenase 1 | 2.44 | 0.025 |
| Oxct1 | 3-oxoacid CoA transferase 1 | 3.57 | 0.035 |
| Protein processing after import | |||
| Hspd1 | Heat shock protein 1 (chaperonin) | 1.47 | 0.025 |
| Hspe1 | Heat shock protein 1 (chaperonin 10) | 2.00 | 0.030 |
| Pmpcb | Peptidase (mitochondrial processing) beta | 2.22 | 0.040 |
| Grpel1 | GrpE-like 1, mitochondrial | 2.70 | 0.043 |
| Others | |||
| Fasn | Fatty acid synthase | −4.80 | 0.030 |
| Dnaja3 | DnaJ heat shock protein family (Hsp40) member A3 | −1.30 | 0.031 |
| Adhfe1 | Alcohol dehydrogenase, iron containing, 1 | 1.52 | 0.030 |
| Prdx5 | Peroxiredoxin 5 | 1.56 | 0.035 |
- Adj., adjusted; CD, control diet; HFD, high-fat diet; TCA, tricarboxylic acid.
Beyond metabolic aspects, we found four proteins involved in processing and folding of nuclear-encoded matrix proteins after mitochondrial import (Table 3) (32). In line with a generally increased matrix protein abundance to cope with, these maintenance proteins were upregulated in response to diet-induced obesity.
Altogether, we demonstrate a general, moderate increase in mitochondrial matrix proteins in subcutaneous adipocytes in response to diet-induced obesity. These proteins mainly constituted key matrix metabolic processes, amino acid catabolism, fatty acid beta-oxidation, and the TCA cycle. Increased requirement of proteins involved in these pathways was associated with increased matrix capacity for protein processing and folding.
Metabolite profiling demonstrates functional consequences of increased matrix metabolism during obesity
The abundance of matrix proteins was increased in adipocyte mitochondria in response to diet-induced obesity. Major pathways affected included beta-oxidation and amino acid degradation. The analysis of acylcarnitines provides information on both pathways. Acylcarnitines represent surrogate measures for the corresponding acyl-CoAs undergoing beta-oxidation and catabolism of several amino acid species (33). We thus analyzed acylcarnitine concentrations in subcutaneous and intra-abdominal AT samples of lean versus diet-induced obese mice. Changes of acylcarnitine concentrations between HFD and CD mice were, in essence, the same in subcutaneous and intra-abdominal AT and differed only in the magnitude of change (Supporting Information Figure S3).
In line with elevated beta-oxidative enzymes (Table 3), we observed an overall increase in fatty acid–derived acylcarnitine species, including medium- and long-chain species decanoyl-, dodecanoyl-, tetradecanoyl-, and hexadecanoylcarnitine (Supporting Information Table S3). Upregulation of enzymes involved in amino acid degradation (Table 3) was accompanied by an overall decrease in intermediates of branched-chain amino acids threonine, methionine, and lysine degradation (methylmalonyl-, succinyl-, and glutaryl-carnitine) (Supporting Information Table S3). Interestingly, whereas the saturated short branched-chain species 2-methylbutyryl-, 3-methylbutyryl-, and isobutyryl- as well as the short nonbranched-chain butyrylcarnitine were decreased, their unsaturated companions such as tiglyl- and crotonylcarnitine were increased. This suggests that, despite an obvious general accumulation of electrons, the reduction reactions between these intermediates might take place. This would require a sustained supply of the coenzyme flavin adenine dinucleotide and liberation of the electrons from the reduced FADH2. In line with this are the increased protein levels of electron transferring flavoprotein (ETF) subunits A and B (Table 3). Furthermore, the increased acylcarnitine levels of hydroxybutyrylcarnitine are indicative of an accumulation of ketone bodies. Thus, a significant increase in matrix enzymes of beta-oxidation and amino acid degradation was accompanied by significant changes in characteristic metabolites, corroborating the notion of altered adipocyte mitochondrial structure and function during obesity.
We further subjected our proteome and metabolite profile data sets to an integrated pathway analysis (Integrated analysis of Cross-platform MicroArray and Pathway data; InCroMAP) (34). This joint analysis identified nine pathways significantly regulated between obese and lean mice, each involving fatty acid metabolism/beta-oxidation, amino acid catabolism, or the TCA cycle (Supporting Information Table S4). Finally, this objective integrated pathway analysis clearly supports our conclusions drawn from the individual data sets, demonstrating significant regulation of typical matrix metabolic pathways during obesity.
Decrease in OXPHOS capacity and cristae density during obesity is a feature of both intra-abdominal and subcutaneous AT
A divergent metabolic risk associated with excessive accumulation of either subcutaneous or intra-abdominal has repeatedly been demonstrated in mice and men (35-39). To detect alterations possibly linked to this difference, we compared obesity-induced changes observed in mitochondria isolated from subcutaneous adipocytes, as reported earlier, with those from intra-abdominal adipocytes.
We observed 55 of a total of 468 mitochondrial proteins to be significantly altered during obesity. The distribution to submitochondrial compartments mirrored that of subcutaneous samples, MIM (61.9%), matrix (23.8%), and MOM (14.3%) (Supporting Information Figure S2). The most prominent effect was downregulation of proteins related to OXPHOS capacity and cristae density (Supporting Information Table S2), again similar to mitochondria from subcutaneous adipocytes. Other functional categories or pathways identified before (MOM: transport; MIM: transport; matrix: fatty acid beta-oxidation, amino acid degradation, TCA cycle, protein processing after import), however, were barely represented and/or inconsistently regulated (Supporting Information Tables S5-S6).
Altogether, we identified systematic downregulation of OXPHOS capacity and cristae density as a general alteration of adipocyte mitochondria in obesity. This held true for both adipocytes of subcutaneous and intra-abdominal fat and was thus independent of anatomical localization. Specifically, mitochondria of subcutaneous, and not intra-abdominal, AT underwent major structural changes accompanied by an increase in matrix metabolic pathways on the proteomic and metabolite level.
Discussion
Expansion of AT is a hallmark of diet-induced obesity. The increase in triacylglyceride storage results in adipocyte hypertrophy and concomitant changes in cellular/mitochondrial metabolism, which have been linked to the development of secondary insulin resistance and type 2 diabetes (1, 14, 15). Previously, we demonstrated limited OXPHOS capacity of adipocyte mitochondria in genetic and diet-induced obesity mouse models (11). Here, we applied an untargeted proteomics approach to determine on the molecular level changes in protein abundance underlying this limitation and to identify novel pathways associated with the metabolic risk emanating from excessive fat mass gain.
Indeed, the key finding of our analysis is an obesity-related profound reduction in OXPHOS enzymatic equipment, accompanied by a moderate decrease in cristae density, observable in adipocytes of both types of AT depots, subcutaneous and intra-abdominal. Albeit very recently, a global decrease in OXPHOS subunits was also found in aging (40), and this level of OXPHOS restriction has not been observed in previous proteome studies on AT in obesity (13, 41, 42). The present results clearly substantiate our previous finding of reduced adipocyte mitochondrial respiratory capacity and impaired mitochondrial functional integrity in adipocytes of obese mice (11). Altogether, profound limitation in adipocyte mitochondrial OXPHOS, the primary function of mitochondria (43), occurs in the obese state on the proteomic, metabolite, and functional level.
Extensive accumulation of intra-abdominal, but not subcutaneous, fat is associated with increased metabolic risk (35, 36, 39). Surprisingly, we detected larger changes in the depot associated with lower risk, that is, subcutaneous AT, and not vice versa. In view of the lower metabolic risk associated with subcutaneous versus visceral fat accumulation, plausibly the structural changes observed in mitochondria of subcutaneous adipocytes in obesity represent a metabolic adaptation and, as such, are of preventive character. Beyond a decreased MIM expanse and OXPHOS complex abundance, these mitochondria feature obesity-induced elevation of matrix enzymes (and metabolites), particularly of matrix enzymes involved in fatty acid beta-oxidation, amino acid catabolism, and the TCA cycle. However, the exact molecular mechanisms conveying this protective action remain elusive at this point. Mitochondria of intra-abdominal adipocytes, on the contrary, fail to undergo these adaptations, thereby increasing metabolic risk associated with visceral fat accumulation.
The functional consequences of the observed mitochondrial reorganization, however, remain elusive. Obesity seems to cause an obvious imbalance between functionally coupled matrix and MIM processes. Increased matrix beta-oxidation, TCA cycle, and amino acid degradation would produce an excessive amount of reduction equivalents that is unmatched by even a reduced mass of OXPHOS machinery. Possibly, amino acid degradation and the TCA cycle are not a priori used to fuel ATP generation but rather contribute to cataplerotic processes. Indeed, amino acids represent a significant carbon source for anabolic processes in differentiated adipocytes (44, 45). In mice fed HFD, the low dietary carbohydrate intake in combination with insulin resistance of adipocytes will limit cytosolic glucose availability in adipocytes. Furthermore, a high dietary fatty acid load demands more glycerol-3-phosphate for esterification of triglycerides. The upregulation of mitochondrial phosphoenolpyruvate carboxykinase (Supporting Information Table S6) indicates upregulated glyceroneogenesis/gluconeogenesis. Amino acids might be used as a carbon source for these pathways by providing succinyl-CoA to the TCA cycle, which is converted to oxaloacetate (Figure 3B). This hypothesis, however, requires further investigation, as, for example, the respective MIM transporters (e.g., Mpc2, Slc25a1, Slc25a10) (Table 2) are not plausibly regulated during obesity.
As a valuable by-product, our study provides information on the applicability of certain proteins for normalization purposes (“housekeeping” proteins). In the field, abundant mitochondrial proteins, including VDAC, HSP60, and central OXPHOS complex subunits, are often used to normalize the mass of other proteins to a “per mitochondrion” base (18, 46-49). We demonstrate that all of these are unsuitable for this purpose when studying adipocytes because they are strongly regulated in response to lipid deposition. Alternatively, our study identifies, for example, the matrix proteins glutathione reductase and superoxide dismutase 2 to be comparable in both feeding groups. Thus, these proteins may be more reliable housekeepers when investigating the effect of obesity on adipocyte mitochondria.
In summary, we describe in unprecedented detail alterations in the mitochondrial proteome of adipocytes in obesity. The key finding, demonstrated for both mitochondria of subcutaneous and intra-abdominal origin, is downregulation of enzymes involved in OXPHOS and cristae density in line with concomitant functional changes demonstrated earlier. This decrease in aerobic ATP production capacity and the underlying structural changes seem to be physiological and beneficial adaptations to the obese state.
Funding agencies
This study was supported by the Else Kröner-Fresenius-Stiftung (Grant 2017_A108-EKFZ) and the German Federal Ministry of Education and Research (BMBF Grant AZ 0315674).
Disclosure
The authors declared no conflict of interest.





