Genome-Wide Study of Subcutaneous and Visceral Adipose Tissue Reveals Novel Sex-Specific Adiposity Loci in Mexican Americans
Funding agencies: This research was supported by DK097524 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), DK053591 from National Institutes of Health, and HG007112 from the National Human Genome Research Institute (NHGRI). The provision of genome-wide association study genotyping data was supported in part by UL1TR000124 (University of California Los Angeles Clinical and Translational Research Institute) and DK063491 (University of California San Diego/University of California Los Angeles NIDDK Diabetes Research Center). Exome chip genotyping was supported by DK081350, HG007112, DK087914, and the Department of Internal Medicine at the University of Michigan. Computational resources were provided, in part, by the Wake Forest School of Medicine Center for Public Health Genomics. The authors would like to acknowledge the members of the GUARDIAN Consortium with research supported by DK085175 from NIDDK and from the Insulin Resistance Atherosclerosis Family Study (IRASFS) grants (HL060944 and HL061019). EKS was supported by NIH grants R01 DK106621, R01 DK107904, The University of Michigan Biological Sciences Scholars Program, and The University of Michigan Department of Internal Medicine. The Multi-ethnic Study of Atherosclerosis (MESA) and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. Funding for SHARe genotyping was provided by NHLBI contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0. Funding support for the abdominal aortic computed tomography data set was provided by grant HL088451.
Disclosure: The authors declared no conflict of interest.
Abstract
Objective
This study aimed to explore the genetic mechanisms of regional fat deposition, which is a strong risk factor for metabolic diseases beyond total adiposity.
Methods
A genome-wide association study of 7,757,139 single-nucleotide polymorphisms (SNPs) in 983 Mexican Americans (nmale = 403; nfemale = 580) from the Insulin Resistance Atherosclerosis Family Study was performed. Association analyses were performed with and without sex stratification for subcutaneous adipose tissue, visceral adipose tissue (VAT), and visceral-subcutaneous ratio (VSR) obtained from computed tomography.
Results
The strongest signal identified was SNP rs2185405 (minor allele frequencies [MAF] = 40%; PVAT = 1.98 × 10−8) with VAT. It is an intronic variant of the GLIS family zinc finger 3 gene (GLIS3). In addition, SNP rs12657394 (MAF = 19%) was associated with VAT in males (Pmale = 2.39×10−8; Pfemale = 2.5 × 10−3). It is located intronically in the serum response factor binding protein 1 gene (SRFBP1). On average, male carriers of the variant had 24.6 cm2 increased VAT compared with noncarriers. Subsequently, genome-wide SNP-sex interaction analysis was performed. SNP rs10913233 (MAF = 14%; Pint = 3.07 × 10−8) in PAPPA2 and rs10923724 (MAF = 38%; Pint = 2.89 × 10−8) upstream of TBX15 were strongly associated with the interaction effect for VSR.
Conclusions
Six loci were identified with genome-wide significant associations with fat deposition and interactive effects. These results provided genetic evidence for a differential basis of fat deposition between genders.
Introduction
Obesity is a global health epidemic affecting more than 500 million individuals worldwide and is responsible for nearly 3 million deaths each year (1). Previous studies have confirmed obesity as a strong risk factor for many metabolic diseases, including cardiovascular disease, type 2 diabetes, metabolic syndrome, certain types of cancer, stroke, and hypertension (2, 3). However, the exact mechanisms underlying these associations have been poorly elucidated.
Recent research has suggested that obesity is not a homogeneous condition, and regional fat distribution affects glucose and lipid metabolism beyond total body adiposity (4). For example, visceral adipose tissue (VAT) has been shown to be responsible for the increased mortality and risk for metabolic disorders, while subcutaneous adipose tissue (SAT) is thought to be benign (5). Genetic studies have been successful in identifying genetic loci responsible for regional fat distribution by using measures including waist circumference and waist-hip ratio (WHR). However, anthropometric measures can be impacted by skeletal structure and aging (6) and cannot differentiate between regional fat depots (e.g., VAT and SAT) and, therefore, bias studies. Currently, computed tomography (CT) is considered as the gold standard for the measurement of adipose tissue deposition (7, 8). However, because of cost and accessibility reasons, only one genome-wide association study (GWAS) has been published focusing on directly measured SAT and VAT with genome-wide significant signals (9).
Numerous evidence has suggested strong sex specificity for regional adipose tissue distributions, with females having a higher proportion of gluteal-femoral body fat, whereas males have more in the abdominal (visceral) region (8, 10). This observation suggests a potentially different mechanism for fat deposition in different genders. In 2015, the Genetic Investigation of Anthropometric Traits (GIANT) consortium published a genetic study of adipose tissue deposition by using WHR and identified 49 (33 new) signals associated with 20 loci demonstrating sex-specific effects (11). Until now, no formal genome-wide single-nucleotide polymorphism (SNP)-sex interaction analysis has been performed in Mexican Americans.
Here, we report a genetic study of sex-specific adipose tissue deposition by using CT measures, including SAT and VAT, in the Insulin Resistance Atherosclerosis Family Study (IRASFS). Genome-wide and exome chip association studies were combined to provide a more comprehensive scan of both common and rare variants. As adiposity deposition differs between genders, sex-stratified analyses as well as genome-wide SNP-sex interaction analyses were performed.
Methods
IRASFS
The study design, recruitment, and phenotyping for IRASFS have been previously described (12, 13). In brief, IRASFS is a family-based study designed to investigate the genetic and environmental basis of insulin resistance and adiposity. Individuals included in this cohort (N = 1,417 individuals; 90 pedigrees) were Mexican Americans recruited from San Antonio, Texas, and San Luis Valley, Colorado. Because a diagnosis of diabetes was not required for participation, about 12.7% of individuals had diabetes. The study protocol was approved by the institutional review board of each participating clinical (UT Health Science Center San Antonio Review Board and Colorado Multiple Institutional Review Board) and analysis (Wake Forest School of Medicine) site, and all participants provided written informed consent.
Phenotypes
Measures of adiposity were obtained by using a standardized protocol. BMI was calculated as weight in kilograms divided by height in meters squared. CT scans were performed to estimate visceral and subcutaneous fat area (VAT and SAT, respectively; centimeters squared). This procedure consisted of a single scout of the abdomen followed by a 10-mm-thick axial image. Axial images were obtained at L4-L5 disc space by using a standard protocol. CT images were sent to a centralized reading center at the University of Colorado Health Sciences Center. VAT and SAT were computed from these data as previously described (7). Visceral-subcutaneous ratio (VSR) was computed as the ratio of VAT and SAT. In addition, glucose homeostasis traits were also obtained in IRASFS (e.g., acute insulin response, metabolic clearance rate of insulin, fasting plasma glucose and insulin, homeostatic model assessment of beta-cell function and insulin resistance). Phenotype acquisition and variable calculations have been previously described (12, 14).
Genotyping and quality control
Genotyping
GWAS genotyping was supported through the Genetics Underlying Diabetes in Hispanics Consortium (15) by using the Illumina OmniExpress and 1S arrays (Illumina Inc., San Diego, California), and exome chip genotyping was carried out on the Illumina HumanExome Array (Illumina Inc.). A detailed description of genotyping platforms and quality controls has been published (13).
Imputation
Imputation was performed by using IMPUTE2 (16) and the 1000 Genomes (1000G) phase I V3 integrated reference panel. All IRASFS samples genotyped on the OmniExpress and 1S arrays were imputed together. Imputed variants were filtered with a confidence score > 0.90 and an information score > 0.50. Imputation quality was evaluated by using 10,000 SNPs with both exome chip and imputation coverage randomly selected from 32,729 overlapping SNPs.
Statistical analysis
GWAS and exome chip
Phenotypes were transformed to approximate the distributional assumptions of normality and homogeneity. Specifically, VSR was natural log transformed, and SAT and VAT were square root transformed. Admixture estimates were calculated by using the maximum likelihood estimation of individual ancestries as implemented in ADMIXTURE (17). For SNPs available in both GWAS imputation and exome chip, exome chip genotypes were always used for analysis. Imputation quality was evaluated with 10,000 overlapping SNPs between GWAS imputation and exome chip, which were selected based on minor allele frequencies (MAF). Concordance analysis was performed between the two platforms and an r squared value was computed for each variant.
Tests of association between individual variants and quantitative traits were computed by using the Wald test from the variance component model implemented in Sequential Oligogenic Linkage Analysis Routines (SOLAR) (SURFsara, Amsterdam, Netherlands) (18). Rare variants (both genotyped and imputed) with MAF < 1% were removed, resulting in a total of 7,708,309 variants. Genetic associations were calculated adjusting for age, recruitment center, admixture estimates, and sex (for sex-combined analysis only). The primary inference was the additive model. A lack of fit to the additive model was tested by using the orthogonal contrast. If the lack-of-fit test was significant (P < 0.05), the model with the “best” P value as the minimum of the dominant, additive, and recessive genetic models was selected. For robust estimation purposes, the dominant and recessive genetic models were not computed if there were less than 10 and 20 individuals homozygous for the minor allele, respectively (this threshold is not applicable for SNPs with the nonsignificant lack-of-fit test). An interaction analysis was performed to test the beta coefficient of the interaction variable by using the same genetic model as the main effect. Genome-wide significance was defined as P < 5 × 10−8, and suggestive significance was defined as P < 5 × 10−7. A genetic locus is defined as a genetic region (< 1 MB) with a cluster of correlated variants (r2 > 0.4). A novel signal is defined as one that is more than 500 kilobase (kb) away from a known CT phenotype-associated locus.
Replication
The replication of loci that attained genome-wide significance in IRASFS was undertaken among Mexican American participants from the Multi-ethnic Study of Atherosclerosis (MESA) (n = 485). MESA is a multiethnic cohort of participants that were free of clinical cardiovascular disease at enrollment (19). Protocols were approved by the institutional review board at each participating institution. All participants provided written informed consent. Assessment of adiposity by CT has been previously described (20). Genotypes from MESA were obtained from the Affymetrix Genome-Wide Human SNP Array 6.0 with imputation to the 1000G phase I V3 integrated reference panel (Thermo Fisher Scientific Inc., Waltham, Massachusetts). The statistical analysis in MESA followed the same protocol as described for IRASFS.
Results
A demographic summary of the study samples is shown in Supporting Information Table S1. Overall, individuals were overweight with an average BMI greater than 28.3 kg/m2. In total, 983 and 1,205 individuals were analyzed for GWAS imputed and exome chip SNPs, respectively. Compared with GWAS, an additional 222 samples were included in the exome chip data, of which 150 were individuals with type 2 diabetes. This resulted in modestly increased age and adiposity traits (P < 0.001). In addition, mean trait values of adiposity phenotypes were significantly different between females and males; i.e., females had significantly larger amounts of SAT (P < 0.0001), while males had larger amounts of VAT (P < 0.0001). The ratio between VAT and SAT (VSR) was two times greater in males than females (P < 0.0001), suggesting males have the predisposition to store fat viscerally. Overall, 7,708,309 SNPs with MAF ≥ 1% were analyzed with SAT, VAT, VSR, and VAT with additional adjustment of BMI (VAT_BMI). A complete list of quantile-quantile plots can be found in Supporting Information Figure S1.
Imputation quality
The majority of the SNPs analyzed were derived from statistical imputation as opposed to direct genotyping. To evaluate imputation quality, a concordance analysis for 10,000 SNPs overlapping between exome chip and imputation was performed. Overall, SNP genotypes were well matched between the two platforms with an r2 > 0.95 (Supporting Information Figure S2). However, for rare variants with MAF < 1%, the imputation quality varied, and therefore, these were excluded from analysis.
Association results
Association analyses were computed for SAT, VAT, VAT_BMI, and VSR, adjusting for age, sex, recruitment center, and admixture estimates. The association results are summarized in Supporting Information Figure S3 and Table 1. The strongest signal identified was SNP rs2185405 (Prec = 1.98 × 10−8; MAF = 40%) with VAT. It is an intronic SNP located in the GLIS family zinc finger 3 gene (GLIS3). The association of this variant in MESA was nonsignificant (P = 0.63; Supporting Information Table S3). In addition, suggestive evidence of the association was observed with SAT (rs2223471 and rs4746598), VAT (rs2131949 and rs78596136), VAT_BMI (rs4243443 and rs12657394), and VSR (rs1504143) (Supporting Information Table S2).
| SNPa | Chr:Pos | Gene | Allelesb | RAFc | β | P overall (N = 983) | β | P female (n = 580) | β | P male (n = 403) | P interaction (N = 983) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VAT | |||||||||||
| rs2185405 | 9:4078851 | GLIS3 | T/C | 0.40 | −0.90 | 1.98E-08d | −0.76 | 1.36E-04d | 1.11 | 1.10E-05d | 0.69d |
| VAT adjusted for BMI (VAT_BMI) | |||||||||||
| rs12657394 | 5:121308199 | SRFBP1 | A/G | 0.19 | 0.62 | 3.68E-07e | 0.35 | 4.10E-03f | 1.09 | 2.39E-08e | 1.10E-03e |
| rs2914610 | 5:121314168 | SRFBP1 | A/G | 0.19 | 0.59 | 9.78E-07e | 0.34 | 5.58E-03f | 1.07 | 4.55E-08e | 1.53E-03e |
| rs1002945 | 7:17796659 | AHR-SNX13 | A/T | 0.43 | 0.50 | 2.80E-05e | 0.10 | 0.33b | 1.09 | 1.17E-08e | 1.37E-04e |
| rs13247968 | 7:17806817 | AHR-SNX13 | G/T | 0.42 | 0.53 | 1.30E-05e | 0.10 | 0.36b | 1.11 | 7.63E-09e | 1.59E-04e |
| rs1830005 | 7:17807563 | AHR-SNX13 | C/T | 0.43 | 0.55 | 6.72E-06e | 0.15 | 0.18b | 1.07 | 3.67E-08e | 8.70E-04e |
| rs9289345 | 3:129579506 | TMCC1 | G/A | 0.01 | 0.35 | 0.51f | 1.40 | 0.017c | −4.59 | 2.90E-04e | 3.73E-08f |
| VSR | |||||||||||
| rs10913233 | 1:176625427 | PAPPA2 | T/A | 0.14 | −0.011 | 0.68f | −0.087 | 4.94E-03f | 0.13 | 4.66E-03e | 3.07E-08f |
| rs10923724 | 1:119546842 | TBX15/WARS2 | C/T | 0.38 | −0.0078 | 0.68f | 0.099 | 2.30E-03e | −0.098 | 3.84E-04f | 2.89E-08f |
- a SNP in build GRCh37/hg19.
- b Minor/major allele.
- c Reference allele based on minor allele.
- d Recessive model; values in bold represent significant results with P < 5 × 10−8.
- e Dominant model; values in bold represent significant results with P < 5 × 10−8.
- f Additive model; values in bold represent significant results with P < 5 × 10−8.
Sex-stratified association analysis
The results of the sex-stratified association analysis are summarized in Supporting Information Figure S4 and Table 1. Overall, five SNPs from two loci reached genome-wide significance (P < 5 × 10−8). Two directly genotyped intronic SNPs (rs12657394; MAF = 18.9%; Pmale = 2.39 × 10−8; Pfemale = 4.10 × 10−3; rs2914610; MAF = 19.2%; Pmale = 4.55 × 10−8; Pfemale = 5.58 × 10−3) within the serum response factor binding protein 1 gene (SRFBP1) on chromosome 5 were strongly associated with VAT_BMI in males (r2 = 0.98). Three imputed SNPs (rs1002945, rs13247968, and rs1830005; r2 > 0.91) located downstream of the sorting nexin 13 gene (SNX13) were significantly associated with VAT_BMI in males but not females (rs13247968; MAF = 42.9%; Pmale = 7.63 × 10−9; Pfemale = 0.36). The analysis of significant results in MESA failed to provide replication (P > 0.35; Supporting Information Table S3), although a consistent direction of effect was observed for the two variants in SRFBP1 (rs12657394 and rs2914610). Signals of suggestive significance (P < 5 × 10−7) are summarized in Supporting Information Table S2.
SNP-sex interaction analysis
Results of the SNP-sex interaction are summarized in Supporting Information Figure S5 and Table 1. SNP rs9289345 was strongly associated with the interaction variable for VAT_BMI (Pint = 3.73 × 10−8; MAF = 1.3%). It is an intronic SNP located within transmembrane and coiled-coil domain family 1 gene (TMCC1). An intronic SNP within pappalysin 2 gene (PAPPA2) (rs10913233; Pint = 3.07 × 10−8; MAF = 13.8%) was associated with the interaction variable for VSR. SNP rs10923724, located upstream of T-box 15 gene (TBX15), was strongly associated with the interaction variable for VSR (Pint = 2.89 × 10−8; MAF = 38.1%). The association of these variants in MESA was nonsignificant (P > 0.15; Supporting Information Table S3).
Assessment of previously identified signals
Nine previously identified CT loci (9, 21, 22) were evaluated herein (Supporting Information Table S4). Consistent with previous findings, the fat mass and obesity-associated gene (FTO) signal was modestly associated with SAT in IRASFS (rs9922619; PSAT = 4.01 × 10−3; Pmale = 5.98 × 10−2; Pfemale = 6.20 × 10−2), yet no interactive effect (PSAT_INT = 0.41) was detected. In addition, SNP rs2123685 on chromosome 7 was modestly associated with SAT and VAT in males (PSAT = 4.15 × 10−2; PVAT = 3.55 × 10−3), and rs7374732 on chromosome 3 was modestly associated with VSR in males (PVSR = 3.21 × 10−2). Furthermore, 49 adipose deposition signals identified by GIANT using WHR were evaluated for association with all four phenotypes (Supporting Information Table S4). Overall, 21 signals were significantly associated (P < 0.05) with at least one of the four CT phenotypes, and 18 of the 21 signals exhibited gender-specific effects (P < 0.05 for gender stratified or interaction analysis). The strongest signal observed was SNP rs2645294 (PVSR_INT = 1.20 × 10−5). It is located in the 3’-UTR region of the tryptophanyl TRNA synthetase 2 mitochondrial gene (WARS2) and has been previously reported to be associated with WHR (11). Interestingly, SNP rs10923724, which is about 500 kb downstream of rs2645294, was identified to have a significant sex-interactive effect (Pint = 2.89 × 10−8) with VSR. Further analysis revealed that the two SNPs were in modest linkage disequilibrium (LD) in IRASFS (r2 = 0.59) and strong LD in Europeans (r2 = 0.93 in 1000G CEU). However, the previous study revealed no sex specificity for SNP rs2645294 (11), suggesting a potentially different biology represented by WHR and VSR.
Discussion
Here we present a study combining genome-wide and exome chip arrays to investigate the genetic determinants of adipose tissue deposition in Mexican Americans from IRASFS. The differentiation between SAT and VAT by using CT provided more refined adiposity phenotypes compared with anthropometric measures. In addition, as SAT and VAT distributions vary by sex, sex-stratified as well as SNP-sex interaction analyses were performed. Signals with significant sex-specific effects were identified.
SNP rs2185405, an intronic SNP within GLIS family zinc finger 3 gene (GLIS3), was found to be genome-wide significant for the main effect with VAT without sex stratification (Pdom = 1.98 × 10−8; MAF = 40%) (Figure 1). On average, the carriers of the minor allele T have 19.91 cm2 less VAT compared with noncarriers. GLIS3 is a member of the GLI-similar zinc finger protein family and encodes a nuclear protein with five C2H2-type zinc finger domains. It functions as both a repressor and activator of transcription and is specifically involved in the development of pancreatic beta-cells, thyroid, eye, liver, and kidney (23). Previous studies have shown that this gene is associated with diabetes in multiple ethnicities (24, 25). In vitro experiments have suggested that GLIS3 modulates pancreatic beta-cell apoptosis via the regulation of a splice variant of the BH3-only protein Bim (26). Because VAT has been shown to be a risk factor for metabolic disorders (4, 5), it is possible that VAT is involved in the modulation of beta-cell function through GLIS3. Further evaluation of the SNP by using glucose homeostasis traits in IRASFS revealed a significant association with insulin clearance (P = 0.03) and a suggestive association with fasting insulin (P = 0.09). However, acute insulin response was not significant (P = 0.64).
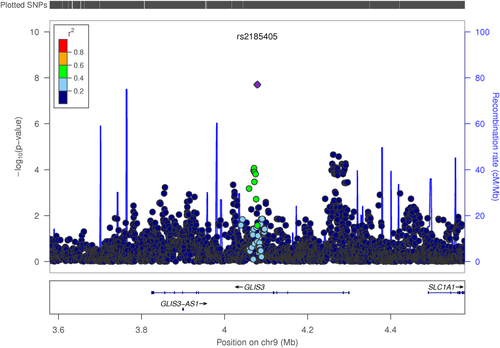
Regional plots of GLIS3 for association with visceral adipose tissue in IRASFS Mexican Americans combining genome-wide and exome chip data sets. [Color figure can be viewed at wileyonlinelibrary.com]
For sex-stratified analyses, five SNPs from two loci (SRFBP1, 7p21.1) reached genome-wide significance. SRFBP1, also named as p49/STRAP, was associated with VAT_BMI in males. Regional plots indicated long-range LD covering a 200-kb region (Figure 2). The strongest signal in the region was an intronic SNP rs12657394 under a dominant model (Pmale = 2.39 × 10−8; Pfemale = 0.0041; MAF = 18.9%). In males, the minor allele carriers had a 24.6 cm2 increase in VAT compared with common allele homozygous individuals. A conditional analysis by using rs12647394 as a covariate abolished the association signal, suggesting only one independent signal exists in SRFBP1. Previous studies have shown that the protein encoded by SRFBP1 specifically interacts with an acidic amino acid motif in the N-terminus of glucose transporter GLUT4 in adipose cells, suggesting a possible role in biosynthesis and/or processing of GLUT4 in adipocytes (27). However, no biological evidence has been identified to explain sex specificity. Further evaluation of glucose homeostasis traits in IRASFS with and without sex stratification revealed modest association signals for rs12657394 with fasting insulin (Padd = 7.92 × 10−3), fasting glucose (Prec = 0.025), homeostatic model assessment of insulin resistance (Padd = 0.03), and homeostatic model assessment of beta-cell function (Pdom = 0.03). However, no sex-specific patterns were observed. An extended examination of this region in the GIANT consortium with waist circumference, BMI, and WHR stratified by sex failed to replicate this signal. This could be due to ethnic heterogeneity as well as the limitations of anthropometric measures compared with CT-derived fat deposition.
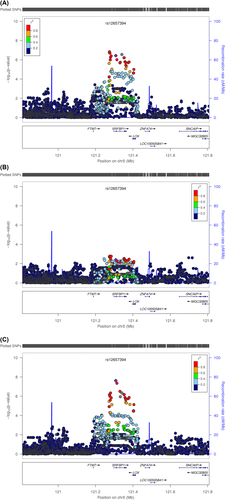
Regional plots of SRFBP1 for association with visceral adipose tissue adjusted for BMI in IRASFS Mexican Americans combining genome-wide and exome chip data sets. (A) Without sex stratification. (B) Females only. (C) Males only. [Color figure can be viewed at wileyonlinelibrary.com]
At 7p21.1, SNP rs13247968 (MAF = 42.5%; Pmale = 7.63 × 10−9; Pfemale = 0.36) as well as two other highly correlated SNPs (r2 ≥ 0.9) were strongly associated with VAT after adjusting for BMI under the dominant model in males. On average, male carriers of the rs13247968 minor allele (G) had a 16.4 cm2 increase in VAT compared with homozygous major allele carriers. The regional plot (Figure 3) revealed a tight cluster of signals located downstream of SNX13 (Sorting nexin 13 gene, also known as RGS-PX1). The RGS (regulator of G protein) domain of the encoded protein can function as a GTPase-activating protein for G alpha subunits of heterotrimeric G proteins, while the PX (Phox) domain works as a sorting nexin protein involved in intracellular trafficking (28). Studies have shown that the protein can target lysosomes and delay lysosomal degradation of the epidermal growth factor receptor (28). Further examination of this region in the GIANT consortium identified that rs1990467, 100 kb proximal to rs13247968, was associated with waist circumference adjusted for BMI in males (P = 2.6 × 10−4) (Figure S6) (29, 30). However, the correlation between rs13247968 and rs1990467 was poor (r2 = 0.00 in IRASFS; r2 < 0.08 in 1000 G). Previous genetic studies have suggested that SNX13 is strongly associated with high-density lipoprotein cholesterol in individuals with European ancestry (31). The evaluation of SNP rs13247968 with circulating cholesterol levels and BMI in IRASFS failed to detect significant associations (P > 0.05). Interestingly, rs13247968 is 300 kb upstream of histone deacetylase 9 gene (HDAC9), which encodes an important histone deacetylase that regulates transcriptional regulation, cell cycle progression, and developmental events. Genetic studies have suggested that HDAC9 is associated with multiple phenotypes, including coronary artery disease, BMI, and vigorous physical activity in European and Hispanic populations (rs2107595, rs2853552, rs12666612, respectively) (32, 33). However, no strong LD was detected between rs13247968 and these previously identified HDAC9 signals (r2 < 0.01).
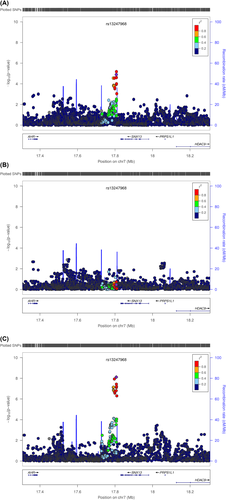
Regional plots of the rs13247968 locus for association with visceral adipose tissue adjusted for BMI in IRASFS Mexican Americans combining genome-wide and exome chip data sets. (A) Without sex stratification. (B) Females only. (C) Males only. [Color figure can be viewed at wileyonlinelibrary.com]
The pappalysin 2 or pregnancy-associated plasma protein A2 gene (PAPPA2) located on chromosome 1 (rs10913233; Padd = 3.07 × 10−8; MAF = 13.8%) was associated with the SNP-sex interaction effect for VSR (Figure 4). The protein encoded by PAPPA2 has been proposed as a biomarker for preeclamptic placenta in pregnant women. It works as a protease, specifically cleaving insulinlike growth factor binding protein 5 (IGFBP5), and thus plays an important role in regulating IGFBP5 levels (34). Diseases related to this gene include hemolysis, elevated liver enzymes, low platelet count (HELLP) syndrome, and developmental dysplasia of hip (35, 36). Human population genetic studies have suggested this gene is associated with height (37). Previous mice studies have identified this gene to be associated with body size and weight, bone size and shape, and postnatal growth retardation, with more pronounced phenotypes in female mice compared with male mice (38). Further examination of this region in the GIANT consortium identified modest signals for WHR and WHR adjusted for BMI in females (rs10913190; PWHR = 5.6 × 10−4; rs10913282; PWHR_BMI = 8.6 × 10−4; r2 < 0.01 with rs12090061 in IRASFS) (Supporting Information Figure S7).
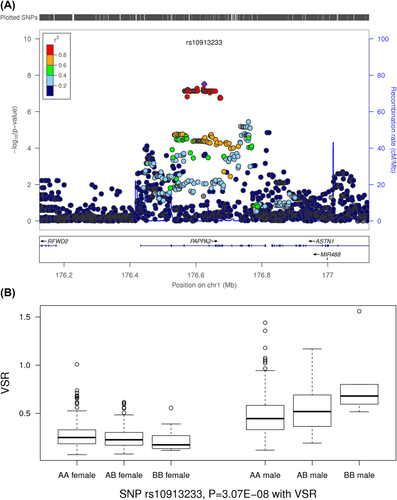
SNP rs10913233 was associated with the SNP-sex interaction variable for VSR. (A) Regional plots of the rs10913233 locus for interaction analysis. (B) Genotypic means of rs10913233 SNP-sex interaction analysis results stratified by sex. [Color figure can be viewed at wileyonlinelibrary.com]
SNP rs10923724 was significantly associated with the SNP-sex interaction variable with VSR (Padd = 2.89 × 10−8; MAF = 38%) (Figure 5). A sex-stratified analysis revealed rs10923724 was nominally associated in males and females with opposite directions of effect (Pmale = 3.84 × 10−4; βmale = −0.97; Pfemale = 2.31 × 10−3; βfemale = 0.10). It is an intergenic SNP located at 1p12, 14 kb upstream of TBX15 and 27 kb downstream of the tryptophanyl tRNA synthetase 2 gene (WARS2). WARS2 is one of the two isoforms of mitochondrial aminoacyl-tRNA synthetases that catalyzes the aminoacylation of transfer RNA (23). The T-box 15 gene (TBX15) is a member of the T-box family. The family encodes phylogenetically conserved transcription factors that regulate developmental processes (23). Diseases related to the gene product include Cousin syndrome and congenital heart malformations (39, 40). Interestingly, this locus has been identified as one of the top WHR genetic signals in multiple ethnicities; rs2645294 (P = 1.7 × 10−19) and rs984222 (P = 8.69 × 10−25) were associated with WHR without sex-specific patterns (11, 41). In IRASFS, the two SNPs were highly correlated (r2 = 0.87) with strong associations with the interaction variable of VSR (rs2645294; PVSR_INT = 1.2 × 10−5; rs984222; PVSR_INT = 2.63 × 10−7). However, they were not associated with noninteractive effects of VSR and WHR (P > 0.10). SNP rs10923724 has nominal correlations with the previous two SNPs (r2 < 0.58) and was not associated with WHR (P = 0.60). An analysis of rs10923724 conditioned by rs2645294 and rs984222 revealed nominal association (P = 0.040) in males and no association in females (P = 0.98). These results suggest there are multiple signals in the region with and without sex-specific heterogeneity. Interestingly, GTEx Portal (Broad Institute, Cambridge, Massachusetts) suggested a strong expression quantitative trail locus signal for rs10923724 with WARS2 expression in multiple tissues, including skeletal muscle and adipose tissue (P = 3.3 × 10−27) (42). Taken together, 1p12 is an interesting locus with complicated signals combining sex-specific and non–sex-specific mechanisms.
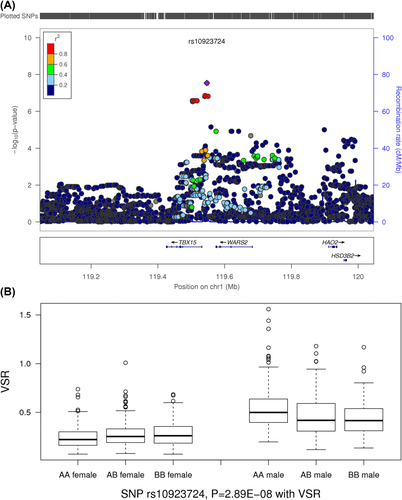
SNP rs10923724 was associated with the SNP-sex interaction variable for VSR. (A) Regional plots of the rs10923724 locus for interaction analysis. (B) Genotypic means of rs10923724 SNP-sex interaction analysis results stratified by sex. [Color figure can be viewed at wileyonlinelibrary.com]
Although significant signals have been identified, study limitations do exist. First, the majority of the SNPs analyzed were from statistical imputation as opposed to direct genotyping. Although SNP genotypes were highly concordant between platforms, the imputation quality was reduced for rare variants with MAF < 1% (Supporting Information Figure S2). Therefore, only SNPs with MAF ≥ 1% were included in analysis. More specifically, the most significant results, with the exception of one, were common variants. Another limitation was the small number of available individuals, which largely limited the study power (i.e., interaction and sex-stratified analyses). The replication efforts focused on Mexican American participants from MESA; however, a modest sample size (n = 485) likely impacted the lack of replication observed. In addition, limited biological evidence was found to support sex-specific signals identified by genetic studies. This is likely attributed to the small number of biological studies that have been focused on sex-specific mechanisms of adipose deposition.
Increasing evidence has supported disease susceptibility heterogeneity related to adipose tissue depots; SAT is benign, while VAT is correlated with metabolic risks (5). The use of CT scans has enabled a more direct estimate of regional adiposity (i.e., differentiate between SAT and VAT). In addition, CT scans are less prone to user bias, as all scans were conducted under the same protocol and results were sent to a centralized reading center. In contrast, anthropometric measures can often be biased by age, sex, clinical site, etc. (6). CT measures can include bias as well. For example, females predominantly store body fat in the gluteal-femoral region, while males store fat in the abdominal region (8). However, CT measures in this study were obtained by axial slices at the L4-L5 disc space, and therefore, no gluteal adipose tissue was measured. This may explain why fewer female signals were observed.
In summary, we computed a combined study of genome-wide and exome chip arrays in the IRASFS Mexican American cohort. Adiposity phenotypes included were SAT, VAT, VSR, and VAT_BMI. Sex stratification and formal SNP-sex interaction analyses were conducted to search for signals with sex-specific effects. These findings support a genetic basis to the differential mechanism of adipose tissue distribution by sex. Moreover, these results highlight the importance of using more refined measures of adiposity (CT scans) as well as the added utility of research in minority populations in which an increased prevalence of adiposity-related diseases may be associated with a different genetic architecture.





