Recommendations for quantitative cerebral perfusion MRI using multi-timepoint arterial spin labeling: Acquisition, quantification, and clinical applications
Abstract
Accurate assessment of cerebral perfusion is vital for understanding the hemodynamic processes involved in various neurological disorders and guiding clinical decision-making. This guidelines article provides a comprehensive overview of quantitative perfusion imaging of the brain using multi-timepoint arterial spin labeling (ASL), along with recommendations for its acquisition and quantification. A major benefit of acquiring ASL data with multiple label durations and/or post-labeling delays (PLDs) is being able to account for the effect of variable arterial transit time (ATT) on quantitative perfusion values and additionally visualize the spatial pattern of ATT itself, providing valuable clinical insights. Although multi-timepoint data can be acquired in the same scan time as single-PLD data with comparable perfusion measurement precision, its acquisition and postprocessing presents challenges beyond single-PLD ASL, impeding widespread adoption. Building upon the 2015 ASL consensus article, this work highlights the protocol distinctions specific to multi-timepoint ASL and provides robust recommendations for acquiring high-quality data. Additionally, we propose an extended quantification model based on the 2015 consensus model and discuss relevant postprocessing options to enhance the analysis of multi-timepoint ASL data. Furthermore, we review the potential clinical applications where multi-timepoint ASL is expected to offer significant benefits. This article is part of a series published by the International Society for Magnetic Resonance in Medicine (ISMRM) Perfusion Study Group, aiming to guide and inspire the advancement and utilization of ASL beyond the scope of the 2015 consensus article.
1 INTRODUCTION
Arterial spin labeling (ASL) perfusion MRI is a non-invasive technique to quantitatively map the supply of blood to an organ. Given the critical role perfusion plays in nutrient delivery, the ability to quantitatively map perfusion has an important role in the diagnosis and monitoring of a wide variety of diseases. The major MRI platforms now all have ASL sequences capable of obtaining high-quality whole-brain perfusion images within a few minutes of scanning. ASL has been validated against exogenous contrast enhanced perfusion MRI1-4 and PET,5-11 has been demonstrated to be reproducible in multi-center studies,12, 13 and has been increasingly deployed in clinical settings.14-19
To achieve simple, robust perfusion quantification, the consensus recommendation, published in 2015,20 was to perform repeated acquisitions using a fixed label duration (LD) and single post-labeling delay (PLD)i; the resulting data being averaged to achieve sufficient SNR for interpretation. An acknowledged limitation of this approach is potential sensitivity to variation in the time it takes for blood to travel from the labeling region to the tissue in each voxel, known as the arterial transit time (ATT). The recommendations controlled for this by recommending a combination of LD and PLD that would be largely insensitive to ATT except where it is prolonged beyond a typical range; formally, single-PLD ASL will be relatively insensitive to ATT when the PLD used is longer than the ATT.23
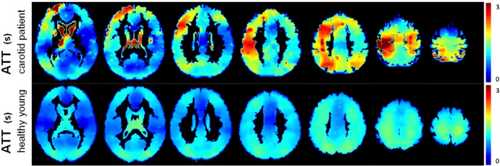
An alternative approach, also discussed in the 2015 consensus paper, is the use of multi-PLD/multi-timepoint ASL.23-28 In this case, the time-based parameters of LD and/or PLD are varied over repeated acquisitions. The resulting data can be combined via kinetic modeling to produce estimates of both cerebral blood flow (CBF) and ATT, with the estimated CBF being inherently insensitive to variation in ATT (see Figure 1).29, 30 This is particularly relevant in settings where the ATT deviates substantially from normal, for example, in steno-occlusive diseases (see Figure 2),33, 34 or is variable between individuals or conditions, for example, in cerebrovascular reactivity (CVR) studies.11 ATT has also been shown to be a useful physiological marker of disease in and of itself.35-44 Finally, multi-timepoint ASL offers the opportunity to extract quantitative maps of additional hemodynamic parameters.45-48
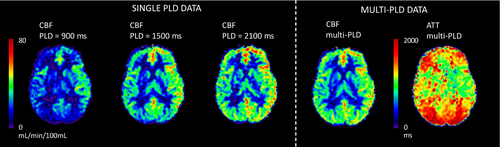
The 2015 single-PLD consensus20 still represents the recommended clinical ASL implementation due to its simplicity and ease of use. However, there has been continued technical development alongside substantial growth in the use of ASL and understanding of how to deploy it in research and clinical practice since that time. Therefore, we believe it is timely to offer specific recommendations for situations where quantitative perfusion metrics derived from multi-timepoint ASL would be preferred and the increase in postprocessing complexity can be accommodated.
2 RECOMMENDATIONS
In this section, we outline the recommendations for multi-timepoint ASL methodology. We build on the 2015 consensus recommendations20 and highlight areas of difference due to the specific considerations when using multi-timepoint ASL. Note: These recommendations have been made for a field strength of 3T and may be sub-optimal at other field strengths. Table 1 provides a concise summary of key acquisition and postprocessing recommendations.
| Key multi-timepoint recommendations |
|---|
| Acquisition |
| 1. Use of pseudo-continuous ASL (PCASL). |
| 2. Protocol design: either sequential, time-encoded and hybrid methods (see Figure 3), depending upon availability and application. |
| 3. Use of variable TRs, where the TR is minimized for each LD/PLD combination, for greater time efficiency. |
| 4. Optimize the background suppression (BS) inversion timings for each TR. |
| 5. Sequence looping: inner-loop is ASL condition (label/control conditions or time-encodings), followed by image segments, then LD/PLD adjustments, with averages in the outermost loop. |
| 6. Scan duration: a protocol of minimum ˜4 min for quantitative multi-timepoint ASL at 3T when reliable parameter estimates are required at an individual level. |
| Postprocessing |
| 7. At minimum, estimate perfusion and ATT from multi-timepoint data using a kinetic signal model. |
| 8. Use of an extended version of the single-PLD ASL quantification model from the 2015 consensus paper that incorporates the effects of variable ATT (Eqs. 2 and 3) |
| 9. In general, model intravascular signals rather than suppress them. |
| 10. When vessel suppression has not been used, include an extra intravascular component in the quantification model (Eq. 6). |
| 11. Use of motion correction to further reduce the impacts of motion beyond what BS can achieve. |
2.1 ASL labeling approaches
Due to its greater SNR, inherent control of the LD, compatibility with modern hardware, and wide-availability, PCASL50 is also recommended for multi-timepoint ASL. Given our recommendation for using PCASL as the labeling technique, many of the following recommendations refer specifically to this technique.
Although pulsed ASL (PASL)51-54 can also be used for multi-timepoint measurements, it has lower SNR. However, when ATT is the primary measurement of interest, rather than CBF, PASL could be considered unless very long ATTs are expected. This is because it is possible to sample more of the label inflow with PASL than with PCASL without reducing the LD. In cases when PASL is used, the QUIPSS II/Q2TIPS21, 55 modification is recommended to control the LD and avoid the need for estimating it during postprocessing. A particular implementation of such methods, in combination with multiple Look-Locker-like readouts, is used in the QUASAR technique.53
Velocity-selective ASL56 is a highly promising alternative to multi-timepoint PCASL/PASL for measuring ATT-insensitive CBF, although in its traditional form cannot measure ATT. The negligible transit time in velocity-selective ASL57 makes it a particularly robust choice for measuring CBF in cases of severely delayed blood arrival.58 A review of velocity-selective ASL and associated recommendations can be found in Ref 22.
2.2 Labeling: spatial placement
An important consideration for multi-timepoint measurements is the PCASL labeling plane location because this has a direct impact on the measured ATT.59
Simple placement options, which only require a fast localizer scan, include placing the labeling plane 85 mm inferior to the anterior commissure-posterior commissure line in adults or just below the inferior border of the cerebellum, although placement between the second and third cervical vertebrae may be more robust across subjects.60 Greater robustness to reduced labeling efficiency in one or more arteries due to tortuous blood flow can be achieved by using an angiographic survey to place the labeling plane. However, because labeling plane placement is particularly robust to angulation of the carotid arteries due to their fast blood flow velocities,61 any additional placement efforts should focus on robust positioning relative to the vertebral arteries. Care should also be taken to avoid placing the labeling plane near dental implants that can lead to large resonance offsets, reducing labeling efficiency.
Within a study, consistency of positioning, robustness within a given workflow, and avoidance of direct tissue saturation effects within the imaging region are key considerations for labeling plane location. It should be noted that positioning is fixed in many commercial implementations. In all cases, we recommend the positioning strategy be reported to allow others to reproduce the protocol.
2.3 Protocol timings
Unlike single-PLD ASL, where multiple volumes are acquired with the same fixed LD and PLD, a multi-timepoint protocol acquires multiple label/control image pairs with a combination of different LDs and/or PLDs. Many methods for acquiring this dynamic data have been developed and, while they can all be successfully utilized, each has advantages and disadvantages which we have summarized here. We recommend that one of the strategies illustrated in Figure 3 is used to achieve a range of LDs and/or PLDs, including sequential, time-encoded and hybrid methods, depending upon availability and application.
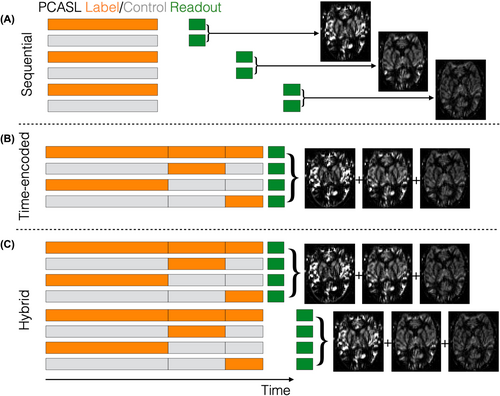
Although multi-timepoint ASL requires the acquisition of more than a single label/control pair, the scan time need not be longer than that of a single-PLD acquisition, where the minimum recommended scan time is ˜2 min20 due to necessary signal averaging. This is typically sufficient to acquire four to five different timepoints, depending on the type of readout used (e.g., single-shot EPI, multi-shot/segmented 3D methods), and although fewer averages can be acquired at each timepoint, the data are combined during the fitting process, thus achieving a similar level of signal averaging and noise suppression to single-PLD ASL but with the advantage of additional hemodynamic information.27, 62
2.3.1 Sequential
The simplest approach to acquire multi-timepoint data is to perform a series of single-PLD scans with varying PLDs.3, 23, 24, 63 However, we recommend that the PLDs are interleaved as much as possible so that the distribution of PLDs acquired at any given time closely matches the final targeted distribution. When each PLD has the same number of averages, this is simply achieved by the sequence looping over each PLD before acquiring the next average. This may reduce motion artifacts by minimizing the acquisition time between each PLD, but also distributes the PLDs throughout the scan, potentially reducing measurement bias from transitory physiological variation. The repeated acquisitions can then be averaged at the end of the scan for visualization. This approach is referred to as sequential multi-timepoint ASL.
Sequential PCASL (Figure 3A) is widely available and may be more robust to subject motion than time-encoded PCASL but, in general, it has lower CBF and ATT precision than hybrid protocols and lower ATT precision than time-encoded protocols (see below).27 Although the LD is often kept constant, it can also be varied with the PLD when using PCASL to provide greater time efficiency57, 64-66 and increased measurement precision.27 Finally, to achieve greater time efficiency, we recommend using variable minimum TRs, where the TR is minimized for each LD/PLD combination in the acquisition (i.e., the time between the end of the readout in one TR and the start of labeling in the next TR is fixed and minimal). When using variable minimum TRs, particular care should be taken to robustly saturate the imaging volume at the start or end of each TR.
2.3.2 Time-encoding
Time-encoded PCASL25, 26, 67, 68 (Figure 3B, sometimes referred to as “Hadamard-encoded PCASL”) was introduced as a time-efficient alternative to sequentially varying the LD and PLD. Rather than acquiring each image with a single combination of LD and PLD, the PCASL preparation is split into multiple sub-boluses. Each sub-bolus has an effective LD and PLD and the acquired images contain a mixture of their signals. The label/control conditions for each sub-bolus are varied across multiple TRs using a linearly independent pattern. Subsequently, signals associated with each sub-bolus can be separated via simple addition and subtraction of the acquired images in a process called “decoding” (see Figure 4).
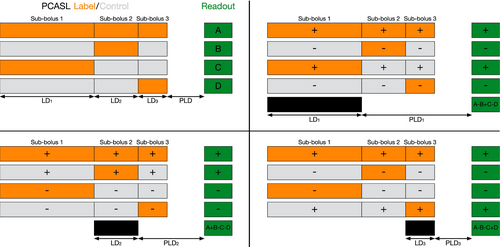
Hadamard encoding69 is typically used to define the encoding pattern because of its optimal efficiency: only N images are required to decode N-1 timepoints and the measurement noise is reduced by a factor of N/2 compared to a matched sequential protocol.25 However, because the labeling period is split into multiple sub-boluses, most of the sub-boluses have relatively short LDs (see Table 2), resulting in lower signal compared to the typically long LDs used in sequential acquisitions. This can decrease, and sometimes outweigh, the noise averaging benefits for CBF accuracy compared to sequential acquisitions, depending on the protocol timings (e.g., see the similar CBF errors for time-encoded and sequential protocols in Figure 5).27, 70 Nevertheless, time-encoded preparations generally have superior ATT accuracy.27, 70
A possible downside of time-encoding is its higher sensitivity to image artifacts, especially those arising from motion, because more than two images must be combined to generate the individual difference images associated with each sub-bolus. Hence, unlike sequential acquisitions, it is not possible to individually remove corrupted label/control pairs. This increased motion sensitivity can be mitigated by using effective BS and Walsh-ordering of the Hadamard matrix.71 Despite this drawback, time-encoding has been successfully used in a range of clinical and research applications.25, 72, 73
2.3.3 Time-encoded/sequential hybrid
A more flexible alternative to pure time-encoding is to sequentially repeat a time-encoded protocol with different final PLDs.27 This hybrid protocol (Figure 3C) enables the use of longer LDs than pure time-encoding because a 4 × 3 Hadamard encoding can be used (smaller than the more typically used 8 × 7 or 12 × 11 encodings), while sufficient temporal information for accurate measurement is generated from both the time-decoding and multiple PLDs (the number of decoded timepoints being three times the number of final PLDs for a 4 × 3 encoding). This approach has been shown to provide more precise CBF measurements than other multi-timepoint protocols27 and the use of a smaller time-encoding matrix reduces motion sensitivity. To further minimize motion sensitivity, the sequence should ideally loop over the encodings, then PLDs, then averages, but a standard time-encoded scan can also simply be repeated with different PLDs.
2.3.4 Look-Locker
Multiple timepoints can also be obtained using a Look-Locker readout,28, 53, 74 where multiple images are acquired with different PLDs after a single ASL preparation. This can be an attractive approach because the time taken to acquire all the PLDs can be reduced compared to other multi-timepoint protocols, making it more robust to subject motion and more suitable for measuring short time-scale perfusion changes. Look-Locker readouts can also be combined with time-encoded PCASL to generate very high temporal resolution data.75
However, there are several limitations of using Look-Locker ASL. First, labeled blood within the imaging volume is attenuated by repeated readout excitations, so low excitation flip angles are necessary to preserve signal for later timepoints,76, 77 which reduces SNR. Second, this attenuation must be accounted for during quantification, which is made difficult due to uncertainty in how much of the bolus was attenuated by each excitation.28, 74, 78 Third, background signal can typically only be suppressed at a single time point, so, as with multi-slice readouts, the effectiveness of BS is reduced (see below). Finally, brain coverage is limited by the time interval between two consecutive measurements (typically five to seven slices are compatible with a 250–300 ms temporal resolution),79, 80 although simultaneous multi-slice (SMS) can be used to increase coverage.75, 81, 82 Due to its reduced SNR and the increased difficulty in its use, Look-Locker ASL is not recommended for routine use if one of the above alternatives is available.
2.3.5 MR fingerprinting ASL
Multi-timepoint data can also be acquired using an MR fingerprinting approach.83-86 In ASL fingerprinting, a series of images is acquired with varied label/control PCASL preparations. Unlike other ASL protocols, the image volume is not saturated between acquisitions, so each image contains contributions from a number of previous label/control preparations. Although an almost zero PLD is typically used, a diverse range of temporal information is encoded by pseudo-randomly varying the LD and label/control order. MRF ASL is still considered to be an experimental method; further details can be found in.87
2.3.6 Optimal timings
The choice of LDs, PLDs, and number of timepoints acquired directly affects the precision and bias of the parameters estimated (typically CBF and ATT as a minimum). In the literature, protocol timings are often chosen empirically. However, timings which maximize measurement precision may also be chosen objectively using the Cramér-Rao lower bound (CRLB), a statistical expression that describes the lower bound of the estimated parameter variances.88
The use of the CRLB for finding optimal multi-timepoint protocol timings has been widely demonstrated for CBF and ATT estimation,27, 28, 62, 74, 83, 89-98 and we recommend that CRLB optimized protocol timings are used for quantitative imaging, when possible, to maximize measurement precision and scan efficiency.
While measurement precision can typically be maximized with many single-average unique timepoints,62 it is often simpler to design an acquisition with a fixed number of unique timepoints and repeat this set as needed to achieve sufficient SNR. In practice four to seven unique timepoints is generally sufficient to estimate CBF and ATT and can be designed to offer superior precision to a single-PLD acquisition.27
Timings, optimized for both CBF and ATT precision, for several classes of PCASL protocol at 3T, covering typical ranges of ATT (0.5–1.8 s and 0.5–2.5 s) and constrained to a 5-min scan duration, are provided in Table 2 with the simulated measurement errors for these protocols, and two single-PLD protocols for comparison, shown in Figure 5. These multi-timepoint protocol timings were generated using freely available open-source software (https://github.com/physimals/oxasl_optpcasl) which readers are encouraged to use if their scan requirements differ substantially from those provided, for example, different field strength (affecting T1b) or incompatible readout segmentation. Note, improved multi-timepoint CBF accuracy can be achieved by optimizing the protocol timings for just CBF precision.62
| Protocol | LDs (ms) | PLDs (ms) | NT | NAve /NSeg | NAcq | Scan duration (min:s) |
|---|---|---|---|---|---|---|
| ATT range 0.5–1.8 s | ||||||
| Sequential Multi-PLD | 2175 | 100, 100, 1275, 1800, 2100 | 5 | 8 | 80 | 5:00 |
| Sequential Multi-LD/PLD | 1925, 1825, 2300, 2300, 2300 | 175, 300, 1300, 1750, 2050 | 5 | 8 | 80 | 5:00 |
| Time-encoded T1-compensated | 1550, 775, 525, 400, 325, 275, 225 | 100 | 7 | 8 | 64 | 4:59 |
| Hybrid T1-compensated | 2250, 925, 575 | 175, 350, 600, 625 | 12 | 4 | 64 | 4:43 |
| ATT range: 0.5–2.5 s | ||||||
| Sequential Multi-PLD | 2050 | 200, 775, 775, 775, 1800, 2275, 2475, 2675, 2800 | 9 | 4 | 72 | 5:00 |
| Sequential Multi-LD/PLD | 2300, 1850, 1825, 1800, 1150, 2300, 2300, 2300, 2300 | 200, 975, 1000, 1025, 1675, 2125, 2375, 2625, 2775 | 9 | 4 | 72 | 4:59 |
| Time-encoded T1-compensated | 2150, 900, 575, 425, 350, 275, 250, 200, 175, 175, 150 | 100 | 11 | 4 | 48 | 4:59 |
| Hybrid T1-compensated | 1800, 850, 550 | 150, 1025, 1350, 1400 | 12 | 4 | 64 | 5:00 |
- Note: When LD/PLD combinations are repeated multiple times, it means these timings are effectively acquired with more averages. For the sequential multi-LD/PLD protocols, each LD is used with the corresponding PLD in the ordered list, i.e., LD1 with PLD1. For the hybrid protocols, the same three encoded LDs are used with each final PLD. Protocol optimization settings: maximum scan duration = 5 min, T1b = 1.65 s, minimum PLD = 100 ms, minimum/maximum LD = 50/2300 ms, 500 ms of non-ASL time (readout, etc), minimum variable TR. The ATT probability was uniform across each ATT range and the ATT distribution was sampled every 10 ms. A linearly decreasing ATT probability extended 300 ms either side of the ATT range to avoid the measurement uncertainty increasing at the ATT distribution edges. Originally optimized for a 75 s scan time to allow time for four readout segments within a 5-min scan.
- Abbreviations: NAcq = total number of acquisitions (one acquisition is one label/control/encoded image); NAve, number of averages; NSeg = number of readout segments; NT, number of timepoints.

As can be seen in Figure 5, the hybrid protocols have the lowest multi-timepoint simulation CBF errors while having similar ATT errors to the time-encoded protocols. The sequential protocols have similar simulation CBF errors to the time-encoded protocols but generally have larger ATT errors. Compared to the single-PLD protocols, which do not provide estimates of ATT, the hybrid protocols achieve lower CBF errors for the majority of the ATT ranges (except for the largest ATTs, which are close in value to the single-PLD value). Although these simulations demonstrate that some multi-timepoint protocols can theoretically provide lower CBF errors than single-PLD ASL, it is currently unclear whether this benefit outweighs the increased postprocessing complexity in clinical settings, unless ATT maps are also required.
While it is possible to measure perfusion when ATTs are longer than 2.5 s, T1-relaxation of the label means that the measurements will have low precision within such short scan durations; longer scan durations or the use of velocity-selective ASL22 may be more appropriate in such cases. The use of longer LDs than those used here (max LD = 2.3 s) can improve CBF precision,27 but long LDs can also increase the influence of tissue T1 on the signal, potentially increasing measurement bias unless tissue T1 is separately measured and included during perfusion quantification (see Section 2.7.4).
2.4 Background suppression
BS is used to improve the SNR of ASL perfusion scans.99-103 Improvements in temporal SNR of 2-3x can be expected,103-105 although the effectiveness of BS with 2D multi-slice and Look-Locker acquisitions is reduced because background signal is typically only optimal for the first PLD.
In multi-timepoint acquisitions, the time between the presaturation and readout often varies with each LD/PLD pair for time-efficiency, for example, when using minimum variable TR. Because the use of identical inversion BS timings for all acquired timepoints results in sub-optimal BS, we recommend that the BS inversion timings are optimized for each TR in a multi-timepoint ASL sequence. These timings are ideally calculated on-the-fly so that users can flexibly change scan timings.100, 101
With PCASL, the BS inversion pulses are often placed during the PLD. However, this is often sub-optimal, particularly for long LD/short PLD combinations, for example, time-encoding. We recommend BS inversion pulses are interleaved with PCASL labeling when necessary to improve BS performance.25, 27, 30, 101, 106, 107 When this occurs, the following adjustments are required: (1) the label/control condition for the remaining labeling period should be switched after each interleaved pulse is applied; (2) the BS inversion pulses should be slab-selective with the inferior edge parallel to and coincident with the PCASL labeling plane to invert already labeled/controlled blood while avoiding perturbing upstream arterial spins. For simplicity, the perfusion signal can be scaled by the net inversion efficiency of the BS inversion pulses in the same way as when they are not interleaved.108
2.5 Vessel suppression
At short PLDs, some labeled blood is likely to remain in the arteries, even among young healthy participants. Intravascular ASL signal can lead to local overestimation of perfusion and underestimation of ATT.30, 109 For accurate quantification of perfusion, intravascular ASL signal should be either modeled or suppressed. In general, we recommend modeling intravascular signals rather than suppressing them (see Section 2.7.1), because they can contain clinically valuable information which vessel suppression can remove110 and vessel suppression typically reduces SNR due to T2 or T2* decay. However, whether including additional intravascular model parameters leads to more or less perfusion measurement uncertainty than using vessel suppression is an unanswered question that requires further research. It is important to note that vessel suppression is commonly applied along a single flow-direction, leading to imperfect suppression.45 Nonetheless, there may be cases when vessel suppression is preferred, such as in tumor imaging,111 making it a valuable option for multi-timepoint ASL.
While the additional ASL signal attenuation from vessel suppression can be modeled, it is simpler and more robust to apply the same vessel suppression during acquisition of the calibration image, thus canceling out this effect after division by the reference image.30
2.6 Readout approaches
Segmented 3D readouts, such as 3D GRASE100, 112, 113 and 3D RARE stack-of-spirals,103, 105 along with judicious use of parallel imaging are recommended for ASL, when available, due to their compatibility with BS, identical PLD across slices, and high SNR.20 With multi-timepoint ASL, the level of readout segmentation possible may be reduced for short scan durations due to the competing use of scan time for acquiring segments and temporal information. A rule of thumb is to only reduce the number of segments if the readout duration can be kept to <300 ms to avoid high levels of through-plane blurring,20 but this will depend on the effective spatial resolution required.
Extending from the 2015 consensus paper, we recommend that the sequence first loops over the ASL condition (label/control conditions or time-encodings), followed by image segments to achieve the most accurate label/control subtractions.20 This should then be followed by looping over LD/PLD adjustments, with averages in the outermost loop.
In some applications, including those where subject motion might be an issue, the use of single-shot 3D acquisitions may be desirable. Recent work has shown that single-shot readout durations can be reduced using in-plane partial Fourier combined with acceleration in the slice direction,114 acceleration along both phase encoding directions115 or employing 3D acceleration.116 Compressed sensing can also be used to exploit the temporal signal redundancy in multi-timepoint ASL by varying the undersampling pattern across timepoints, enabling increased acceleration factors.66
2D single-shot readouts (e.g., EPI and spiral) are an adequate alternative when an optimized 3D readout is not available, although BS effectiveness is reduced and the PLD varies across slices, leading to slice-dependent CBF and ATT accuracy.62 SMS can increase the number of slices to achieve whole brain coverage,117, 118 improve BS119 and reduce PLD differences across slices. However, the performance of postprocessing motion correction strategies can be compromised.120
2.7 Postprocessing methods
Whilst it is important for quality assurance to examine the ASL difference (control-label) images, we recommend that multi-timepoint ASL is processed using a quantification model to arrive at estimates of perfusion and ATT as a minimum. A major benefit of multi-timepoint ASL is being able to account for the effect of variable ATT on quantitative perfusion values and visualize the pattern of ATT itself. Since the tracer kinetics of the ASL label results in a signal model that is non-linear in ATT, it is not possible to use a single formula to compute perfusion, as was provided in the 2015 consensus paper. Instead, it is necessary to perform some form of algorithmic analysis. Although this is more complex to implement, various existing algorithms and software tools are now readily available for this task (see: OSIPI (Open Science Initiative for Perfusion Imaging) pipeline inventory).121
2.7.1 Quantification model
The General Kinetic Model (GKM) is the most universal and widely used signal model for ASL. It was outlined in detail by Buxton et al.29 following principles used in the early ASL publications52, 122-124 and based on indicator-dilution theory.125 By describing the delivery of labeled blood water to a voxel via an “arterial input function” and the subsequent behavior of that labeled blood water after arrival, via the “residue function,” the GKM provides a flexible way to incorporate a range of effects on the ASL signal.
In the 2015 consensus paper, several assumptions were made to derive a simplified model for perfusion quantification from single-PLD ASL based on the GKM. This was chosen as it can be used robustly in a wide range of circumstances without the need for more than a few key parameters to be set. We recommend that, in general, an extended version of this model is used for multi-timepoint data that incorporates the effects of variable ATT, but otherwise retains the other assumptions. This can be described by the following equations for PCASL and PASL:
where and are respectively the signal intensities in a single pair of control/label images acquired with the same LD and PLD, α is the labeling efficiency, αBS is the total BS inversion efficiency, T1b is the longitudinal relaxation time of arterial blood in seconds, and M0a is the equilibrium magnetization of arterial blood, calculated as ,126-128 where SIPD is a proton density weighted image and λ is the tissue/blood partition coefficient (see Ref 128 for a detailed discussion of M0a calibration approaches). The factor of 6000 converts the units for CBF from mL/g/s to mL/100 g/min. Note that for 2D multi-slice imaging, the PLD value should be adjusted for each slice to account for the time delay between slice acquisitions.
The major assumptions of this model are consistent with the recommended single-PLD formula, that is, (1) there is no outflow of labeled blood water and (2) the relaxation of labeled spins is governed by blood T1 (or equivalently that blood T1 and tissue T1 are equal).20 The second assumption may introduce appreciable errors where differences in T1 between blood and tissue are large (e.g., in white matter [WM] and tumors) or when measurement times (LD + PLD) are long.129 More complex models and separate measurements may be used to mitigate this, as discussed below.
When vessel suppression has not been used, we recommend that an extra intravascular component is included in the quantification model. The models in equations Eqs. (2) and (3) assume that all labeled blood water has arrived in the capillary bed at the PLD, that is, it is a result of perfusive delivery to the tissue. Signal contributions arise from labeled blood water that remains in arterial vessels at the time of imaging: the so called “intravascular signal.” For the purposes of CBF quantification, this signal is regarded as artifactual, giving rise to overestimation of perfusion in voxels displaying intravascular contamination as the labeled blood water in the vessels is destined for delivery to tissue in other voxels via downstream capillary vessels. Since this signal is typically associated with larger arterial vessels with high flow speed compared to the rate of water exchange across the vessel walls, we recommend that this macrovascular component is modeled with the same form as the arterial input function.45 Using similar assumptions as above, this gives rise to the equation:
2.7.2 Model fitting
For multi-timepoint ASL, data at different timepoints need to be combined and multiple parameters estimated. Various methods have been used to estimate kinetic model parameters in the literature, but the majority can be viewed as some form of nonlinear regression. There are examples of semi-parametric approaches being used to estimate a surrogate of the ATT from features of the multi-timepoint time course and using this ATT surrogate in the calculation of CBF.30, 53, 130 Alternatively, the GKM may be fit directly to the data for CBF and ATT using a nonlinear model fitting algorithm such as non-linear least squares29 or Bayesian inference.131 The advantage of a full model-based fitting is the ability to flexibly extend the model and fit more parameters, for example, themacrovascular component, where the data allows. There are few studies directly comparing different ASL analysis algorithms,39, 132 although the Open Science Initiative for Perfusion Imaging has developed inventories of ASL software tools121 and challenges133 to make systematic comparisons (www.osipi.org). There is currently no strong evidence to favor one algorithm over another in terms of accuracy, although features, for example, ability to model effects such as macrovascular contamination, spatial regularization, or uncertainty estimation might favor particular algorithms in specific cases.
Kinetic model analysis has been combined with the image reconstruction process to enable greater levels of acquisition acceleration by utilizing additional temporal model based sparsity.66 In contrast to non-linear regression algorithms, MRF ASL has tended to employ dictionary algorithms (based on those used in the early MR fingerprinting literature).85, 86 These are still based on a kinetic model description of the signal, but precompute various examples of the timecourse corresponding to the acquisition scheme used and find the closest match to the data. These algorithms can offer fast processing and robust performance, especially when a more complex model is adopted, but can suffer from limitations due to the size of the precomputed dictionary and only being able to select from a discretized set of the parameter space. There are a growing number of examples of machine and deep learning analyses for MRI data, including ASL.83, 134-136 For the most part these currently seek to learn either an existing kinetic model-based analysis or the relationship to another perfusion imaging measure. Such methods can offer fast analysis and robust solutions, but currently need to be trained for a specific acquisition protocol and do not generalize readily.
2.7.3 Variation in blood T1
Blood T1 has a global scaling effect on the estimated perfusion and its value can be fixed according to literature values, for example, 1650 ms for 3T.137 Experimental measurement of subject-specific blood T1, either directly138-141 or using non-imaging parameters with a physiological model,137, 142-144 can also be considered when blood T1 deviates significantly from the normal range, such as in anemia145 or gas challenges.146 The value of obtaining per subject measures of blood T1 will depend on the application, but there is some evidence that the reduction in perfusion bias from spending some of the available scan time measuring blood T1 might outweigh the loss in precision for the ASL scan in terms of overall accuracy.147
2.7.4 Variation in tissue T1
More complex models can mitigate the assumption that the relaxation of the labeled spins is entirely governed by blood T1, accounting for the time the labeled blood water spends in the tissue. This is particularly important for non-cortical brain regions with shorter T1 values such as WM, deep gray matter (GM) structures with high iron content such as the pallidum and putamen, and in tumors or other pathology where T1 may be shortened.148 In these situations, the single-compartment model of Buxton et al.29 can be used with independent values for blood and tissue T1, which can either be set using tissue specific literature values149, 150 or measured using a separate scan. In cases where the tissue T1 does not deviate greatly from typical values, it is unclear if the reduced perfusion bias from separately measuring tissue T1 outweighs the loss in perfusion precision due to spending less of the available scan time on the ASL acquisition.
Although the tissue T1 can be estimated from the ASL data itself during the quantification process (i.e. simultaneously with CBF and ATT),63 this decreases the precision and repeatability of the perfusion measurements, outweighing the benefits of reduced measurement bias for typical spatial resolutions and scan times.89 Some ASL techniques,65 including fingerprinting,84-86, 134, 136 use a saturation-recovery approach to provide another source of tissue T1 information in the ASL data that is not coupled to the perfusion signal; however, this generally precludes the use of efficient BS.
Use of the model with separate blood and tissue T1 introduces an additional assumption that labeled blood water exchanges instantaneously with tissue water as soon as it enters the tissue voxel (i.e., when LD + PLD > ATT). In practice, labeled spins will remain in the blood compartment for some time prior to exchange. More complex two-compartment models incorporating exchange time or permeability of the vessel wall to water can mitigate this assumption.129, 151 However, knowledge of water exchange is then needed which is challenging and time-consuming to measure accurately.152-154
2.7.5 Bolus dispersion
In the standard ASL kinetic model, labeled blood moves downstream inside arteries under the assumption of a uniform velocity profile (plug flow). In reality, the bolus of labeled blood water may be subject to intravascular dispersion. Dispersion affects the temporal features of the intravascular ASL signal and consequently the estimated perfusion value through changes in the form of the arterial input function. In a single-PLD ASL experiment, it is not possible to reconcile these issues and, therefore, dispersion contributes to errors in perfusion estimation. Characterizing ASL dispersion itself may be of interest since the process relates to cardiovascular physiology. Pathological changes to the composition of arteries can increase arterial stiffness, altering the degree of ASL bolus dispersion.47 Similarly, narrowing of the arterial lumen can alter the fluid mechanics inside the conduit ASL vessel.155 However, few studies have investigated ASL dispersion in relation to the underlying physiological drivers.
More advanced models have been used to characterize dispersion, such as modeling dispersion using a vascular transport kernel47, 153-157 or using typical intravascular flow velocity characteristics to describe the expected form of the arterial input.158 Multi-timepoint measurements offer the possibility to detect variation in the signal due to dispersion and attempt to correct for it by inclusion of further dispersion parameters in the model fitting, potentially yielding further hemodynamic information in the process. The effects of dispersion are most noticeable on signal from arterial vessels and this has been used to characterize dispersion in ASL using angiographic readouts.47, 159, 160 The effects of dispersion on perfusion estimates can be substantial,160-162 but can be hard to detect in the signal time course because dispersion effects can appear similar to variations in other model parameters. Consequently, fitting dispersion parameters accurately from ASL tissue signal is challenging unless high SNR and high temporal resolution data is available.160
The most widely used model of dispersion thus far is the gamma vascular transport kernel,47 which can be convolved with the above models. This provides a good fit to ASL data and is relatively mathematically simple, adding only two additional model parameters. To avoid the risk of overfitting when implemented in a model fitting algorithm, the dispersion parameters should be constrained to a realistic range, e.g., using a Bayesian prior, or fixed to literature values; for example, Ref 47 found values for the gamma vascular transport function of s ˜ 0.5 and p ˜ 0.1 in major arteries. Where a macrovascular component is present and included in the model, this may also be used to simultaneously estimate appropriate dispersion parameters for the dataset.
2.7.6 Partial volume effects
Differences in perfusion demands between tissues, combined with the low spatial resolution of ASL images relative to anatomical tissue variation, leads to a partial volume effect (PVE): the voxelwise measured CBF is a tissue-weighted average measure.46 Correcting for PVE is especially relevant in studies of aging and dementia because PVE is exacerbated by atrophy. Algorithms that correct PVE (PVEc) typically use PV information derived from a high-resolution anatomical scan.31, 163 Given that this scan is already routinely acquired in imaging studies, we encourage PVEc to be performed as an additional analysis, especially for clinical studies and studies where structural and hemodynamic changes may co-occur.
Since each tissue compartment in the brain has different perfusion kinetics (WM typically has longer ATTs than GM), multi-timepoint ASL is particularly suitable for PVEc since it provides extra information for a PVEc algorithm to separate different tissue contributions.163 PVEc with multi-timepoint ASL not only provides separate estimates of GM and WM perfusion but also ATT for both tissue components.
2.7.7 Motion correction
Subtraction of label/control images is an essential step in ASL postprocessing164 but makes ASL especially sensitive to motion. Subject motion should, therefore, be minimized as much as possible, which is typically achieved with foam pads. While BS reduces the impacts of motion, in general, we recommend the use of motion correction as another important strategy. This can take the form of prospective motion correction,165, 166 motion correction during image reconstruction,167-170 or image-based registration during postprocessing. Motion-correction of the unsubtracted image series using rigid-body transformations during postprocessing is most commonly used due to its wide availability. However, this type of motion correction can be challenging for data with efficient BS because there is little static tissue signal available.
It should be noted that, for multi-timepoint data, the effectiveness of BS can differ across the different PLDs, thereby leading to varying image intensities and contrasts of the unsubtracted static tissue across timepoints. This can lead to minor artifactual motion estimations across timepoints when using conventional image similarity-based motion correction algorithms,171 in a similar way to the varying image intensities of label and control images.172 Additional subtraction artifacts and challenges can also arise for motion correction when using simultaneous multi-slice readouts due to the abrupt changes in image intensities across slices.120
2.8 Multi-timepoint ASL in application
2.8.1 Acquisition duration
As noted by the 2015 consensus, since the ASL perfusion signal is small, ASL relies on averaging to achieve sufficient SNR. For multi-timepoint ASL, as a minimum, a ˜2-min protocol including 5 different LD/PLD combinations may be sufficient to acquire quantitative parameter estimates. However, in general, a protocol of minimum ˜4 min is recommended for quantitative multi-timepoint ASL at 3T with the recommended spatial resolution (3–4 mm in-plane, 4–8 mm through-plane)20 when reliable parameter estimates are required at an individual level.
2.8.2 Quality assurance
- Check the temporal dynamics. Scrolling through the different ASL timepoints is the most direct method to identify transitory artifacts or unexpected dynamics (such as arteriovenous malformations [AVMs]), which may be less clear in the final CBF and ATT maps. This can be achieved by first averaging the repeated acquisitions at each timepoint after the control-label subtraction/decoding, then ordering them with increasing TI (LD + PLD). Inspecting the individual difference images can be a useful follow-up step to check the proportion of images at a given timepoint affected. Note, if both the LDs and PLDs are varied, it may be more complicated to manually interpret the signal dynamics.
- Compare the quantified CBF and ATT maps. It is possible that regions that have both low CBF and very long ATTs (where the ATTs are equal to or greater than the longest PLDs used) may in fact be well perfused, but the longest PLDs used were too short to capture the arrival of the ASL label.
- For PCASL scans, look for areas of low labeling efficiency.
- Note the overall GM perfusion value.
- Check for motion artifacts.
- Look for intravascular artifacts.
- Check the border-zone (watershed) regions for artifactually low perfusion which may be due to long ATTs (see new QA steps above).
2.8.3 WM perfusion
Measurement of WM perfusion, and hence detection of WM perfusion abnormalities, is challenging due to low SNR caused by the lower blood flow and longer ATT of WM compared to GM.90, 173-177 Partial voluming with GM may also mask WM perfusion signals in GM/WM border regions.175 The earlier timepoints often used by multi-timepoint ASL strategies may not contribute to greater WM perfusion SNR when compared to the use of a single long PLD. Hence, where WM perfusion is specifically of interest, it may be beneficial to design the timepoints used to include longer ATT or specifically only for a range of ATT seen in WM. Acquisition at a lower spatial resolution,177 or analysis which combines voxels into lower resolution elements, can increase SNR and PVEc can be used to separate WM signals.
3 CLINICAL APPLICATIONS
In this section, we examine examples where multi-timepoint ASL may offer advantages over the existing consensus single-PLD approach.
3.1 Cerebrovascular reactivity
CVR reflects the capacity of blood vessels to alter their caliber and thus modify CBF in response to a vasoactive stimulus, e.g., acetazolamide, CO2, or breath-hold.178-180 This parameter has been shown to be impaired in pathologies where the cerebrovasculature is compromised, such as stroke,181, 182 small vessel disease,183, 184 glioma,185-187 and neurodegenerative diseases.188-191
ASL has been increasingly used to map CVR due to its ability to provide non-invasive and quantitative measures of CBF changes, compared to indirect or invasive methods such as BOLD-FMRI or PET. However, the application of vasoactive stimuli can lead to changes in ATT (see Figure 6),192, 193 potentially compromising CVR measurements when single-PLD ASL is used. Due to its robustness to changes in ATT, multi-timepoint ASL CVR measurements may, therefore, be more accurate than single-PLD ASL.11, 194 However, the short duration of breath-holds can make the use of multi-timepoint ASL difficult for this type of stimulus.180
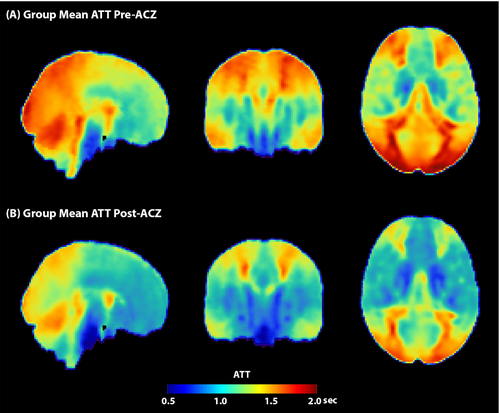
3.2 Steno-occlusive disease
In steno-occlusive diseases, significant transit delay can occur distal to the stenosis, resulting in elongated ATTs and associated transit artifacts when using single-PLD ASL. As the degree of stenosis may vary with disease severity and with underlying etiology, simply choosing a single longer PLD may not be the best approach, making multi-PLD ASL highly relevant for these patients. Multi-PLD ASL has been utilized in steno-occlusive disease patients in both research and clinical settings, primarily in moyamoya disease195, 196 but also in intracranial atherosclerotic disease.197 In one cohort of patients with moyamoya, CBF derived using multi-PLD ASL showed a larger effect size than CBF from single-PLD ASL when comparing pre- and post-revascularization data.198 In the same study, the additional ATT information from multi-PLD ASL was shown to be correlated with dynamic susceptibility contrast-based time-to-maximum values.198 These data show the promise of multi-PLD ASL to provide similar information without the need for contrast injection. Another study in moyamoya patients demonstrated that hemodynamic parameters derived from multi-PLD ASL exhibited better agreement with analogous measures derived using oxygen-15 positron emission tomography than single-PLD ASL in both diseased and healthy brain hemispheres.199 Together, these results support the use of multi-PLD over single-PLD ASL in patients with arterial steno-occlusive diseases.
3.3 Arteriovenous malformations and fistulas
Cerebral AVMs and fistulas characteristically demonstrate abnormal arteriovenous connections (so-called shunts) that allow the ASL signal to bypass the capillary network, resulting in early venous arrival. This unique property of arteriovenous shunts (AVS) typically produces easily identifiable hyperintense venous ASL signal,122, 200, 201 although the degree of conspicuity is dependent on the LD and PLD.
While single-PLD ASL data, acquired using the 2015 consensus parameters, are often useful for assessing AVS, sensitivity may be degraded by several factors. First, single-PLD methods may miss or sub-optimally characterize lesions if shunt flow is too fast (resulting in complete venous clearance) or too slow (resulting in insufficient accumulation). Second, ambiguity will arise when it is unclear if the hyperintense ASL signal truly localizes to a vein versus a non-venous structure (e.g., a nearby artery or abnormal vascular tissue).
The qualitative use of multi-timepoint ASL addresses both issues. Short-PLD images improve assessment of high-flow shunts, whereas long-PLD images improve assessment of low-flow shunts. Since AVS are quite heterogeneous across the patient population, use of multi-timepoint ASL could effectively widen the operating range for ASL. Additionally, multi-timepoint ASL could allow a more comprehensive assessment of shunt flow by permitting a dynamic evaluation that would help spatially localize arterial supply and venous drainage. This type of evaluation is particularly useful when following AVS over time to determine progression and/or response to therapy.
3.4 Aging and neurodegenerative disease
ATTs have been shown to increase with age.130, 202 Therefore, single-PLD ASL may not be well-suited for quantifying CBF in aging populations due to the large variability in the appropriate single-PLD. In these cases, multi-timepoint ASL may be appropriate to enable more accurate quantification of CBF in older adults and has recently been included in large consortium studies involving aging populations, including the Human Connectome Project-Aging,203 UK Biobank, and the Alzheimer's Disease Neuroimaging Initiative.204 Recent work has demonstrated that CBF measured using multi-timepoint ASL exhibited better agreement with CBF from 15O-PET and CT than single-PLD ASL in patients with pathologically long ATTs,6, 195 and that it reduces the variability of CBF measurements in the frontal, parietal, and occipital brain regions in older adults.202 Multi-timepoint ASL has also detected unique patterns in regional CBF in individuals who exhibited variable longitudinal changes in cognitive function.205
In addition to aiding in more accurate quantification of CBF, recent studies suggest that ATT can be used as a cerebrovascular biomarker in aging and neurodegenerative diseases. One study showed that ATT was increased in several regions of interest (ROIs) in patients with Alzheimer's disease compared to healthy controls; and that this ATT increase was associated with cognitive performance.206 Parkinson's dementia207 and multiple sclerosis208 have also shown associations between ATT and cognitive decline. ATT proxies, such as FEAST-ATT209 and the spatial coefficient of variation (sCoV),210 have also been used as potential biomarkers in neurodegenerative disease.
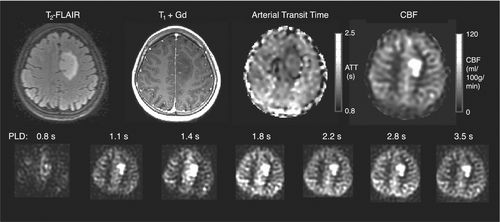
3.5 Cancer
ASL is increasingly used clinically to aid the initial diagnosis of brain tumors and to monitor post-treatment to differentiate tumor recurrence from treatment effects.211, 212 Although multi-timepoint ASL can provide greater CBF accuracy, brain tumor imaging may mostly benefit from its ability to characterize the arterial signal component and aBAT/ATT, due to the common irregular vasculature found with this disease (see Figure 7). For example, the vascular-weighted signal from early (<500 ms) PASL TIs have been shown to be superior to longer TIs in distinguishing between low- and high-grade astrocytoma and glioblastoma, improving pre-operative grading.213 A further study demonstrated that multi-timepoint CBF could distinguish between grade 2, 3, and 4 astrocytoma, whereas single-PLD CBF was only able to distinguish grade 2 and 4 tumors.214 Another study found that, although multi-timepoint PASL with and without vascular crushing was not able to distinguish between different pediatric brain tumors (which DSC was able to), an improved picture was given of the tumor macro- and microvascular compartments.111 The authors of the latter study also indicated that specifically in enhancing tumors, where DSC measurements are affected by leaky vessels, multi-timepoint ASL can likely improve knowledge on perfusion due to the ability to model ATT and intravascular signal. Future studies including multi-timepoint ASL in oncology are, therefore, warranted to further elucidate tumor hemodynamics.
4 SUMMARY
ASL is now an established non-invasive technique for quantitatively imaging cerebral perfusion in both research and clinical settings. The recommendations provided in this paper aim to serve as a comprehensive guide for clinicians and researchers navigating the large array of options available when pursuing quantitative accuracy or the estimation of additional hemodynamic measures using multi-timepoint ASL. By employing a robust, yet relatively simple, multi-timepoint protocol, in conjunction with widely available postprocessing techniques, more accurate and precise measurements of CBF can be achieved by accounting for variable ATTs and intravascular signal, as well as providing clinically valuable measurements of these physiological parameters. These guidelines do not intend to supersede the recommendations outlined in the 2015 consensus paper; rather, they offer specific guidance for utilizing multi-timepoint ASL methods that are already accessible to the research and clinical community through various vendor implementations and software toolboxes.
ACKNOWLEDGMENTS
The work in this article has been endorsed by the ISMRM perfusion study section and by the ISMRM's board of trustees. Endorsement was obtained by a survey among the perfusion study group's members and by subsequent endorsement by the board of trustees.
J.G.W. and T.W.O. were supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (220204/Z/20/Z). J.G.W. acknowledges Linacre College (Oxford) for their support. The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z). A.P.F. was supported by the National Institute of Neurological Disorders and Stroke (R00-NS-102884). J.G. was supported by US National Institutes of Health (R01EB033210). MRJ acknowledges funding from the US National Institutes of Health (K01-AG070318). A.J.M. acknowledges funding from the US National Institutes of Health/National Cancer Institute, U01 CA207091. M.J.Pv.O. acknowledges funding from the Dutch Research Foundation, VICI-project 016.160.351. J.P. was supported by the Engineering and Physical Sciences Research Council (EPSRC) grant EP/S021507/1. M.F.S. acknowledges funding from the Spanish Ministry of Science and Innovation (PI21/00578). D.L.T. was supported by the UCLH NIHR Biomedical Research Centre and the Wellcome Trust (Centre award 539208). G.Z. acknowledges funding from the US National Institutes of Health (R01-EB025220 & R01-NS123025).
Authors are listed in alphabetical order except for the first and last authors. For the purpose of open access, the authors have applied a CC BY public copyright license to any author-accepted manuscript version arising from this submission.
CONFLICT OF INTEREST STATEMENT
D.B. received research grant support from GE Healthcare. M.A.C. receives royalties from the commercial licensing of FSL and is an employee of, and holds equity in, Quantified Imaging Ltd. X.G. is a founder, shareholder and employee of Gold Standard Phantoms and a consultant at Bioxydyn. T.O. works as an ad hoc consultant for SBGNeuro Ltd. M.J.Pv.O. receives research support from Philips. M.S. received consultation fees from Bracco and a speaker fee from Instituzione Internazionale Menarini (both paid to Erasmus MC). D.J.J.W. is a shareholder of Hura Imaging, INC. G.Z. receives grant support from GE Healthcare and Bayer Healthcare and has equity interest in Subtle Medical, Inc.
ENDNOTE
Open Research
DATA AVAILABILITY STATEMENT
The MATLAB code used to perform the Monte Carlo simulations in Figure 5 are available at https://doi.org/10.5281/zenodo.10642060.




