Compensation for respiratory motion–induced signal loss and phase corruption in free-breathing self-navigated cine DENSE using deep learning
Click here for author-reader discussions
Funding information: National Institutes of Health, Grant/Award Numbers: R01HL147104, R01HL159945
Abstract
Purpose
To introduce a model that describes the effects of rigid translation due to respiratory motion in displacement encoding with stimulated echoes (DENSE) and to use the model to develop a deep convolutional neural network to aid in first-order respiratory motion compensation for self-navigated free-breathing cine DENSE of the heart.
Methods
The motion model includes conventional position shifts of magnetization and further describes the phase shift of the stimulated echo due to breathing. These image-domain effects correspond to linear and constant phase errors, respectively, in k-space. The model was validated using phantom experiments and Bloch-equation simulations and was used along with the simulation of respiratory motion to generate synthetic images with phase-shift artifacts to train a U-Net, DENSE-RESP-NET, to perform motion correction. DENSE-RESP-NET-corrected self-navigated free-breathing DENSE was evaluated in human subjects through comparisons with signal averaging, uncorrected self-navigated free-breathing DENSE, and breath-hold DENSE.
Results
Phantom experiments and Bloch-equation simulations showed that breathing-induced constant phase errors in segmented DENSE leads to signal loss in magnitude images and phase corruption in phase images of the stimulated echo, and that these artifacts can be corrected using the known respiratory motion and the model. For self-navigated free-breathing DENSE where the respiratory motion is not known, DENSE-RESP-NET corrected the signal loss and phase-corruption artifacts and provided reliable strain measurements for systolic and diastolic parameters.
Conclusion
DENSE-RESP-NET is an effective method to correct for breathing-associated constant phase errors. DENSE-RESP-NET used in concert with self-navigation methods provides reliable free-breathing DENSE myocardial strain measurement.
1 INTRODUCTION
Cine displacement encoding with stimulated echoes (DENSE) is an accurate and reproducible method of myocardial strain imaging and provides automatic displacement and strain analysis.1-3 These properties have led to increasing clinical applications. For example, Bilchick et al. showed an important role of DENSE for prognostication in heart failure patients undergoing cardiac resynchronization therapy,4, 5 Mangion et al. showed the prognostic value of DENSE in acute myocardial infarction,6 and Jing et al. showed that DENSE detects systolic dysfunction in childhood obesity.7 While these studies used breath-hold or navigator-based DENSE protocols, breath-holding is taxing for patients, and diaphragm navigator methods are burdensome for technologists, prompting the need for self-navigated free-breathing DENSE.8
The various echoes generated by a DENSE pulse sequence have been described previously.9-11 In free-breathing DENSE, respiratory motion can lead to three types of artifacts. The first type is image striping due to imperfect suppression of the T1-relaxation echo.8 The second type is blurring due to motion-induced position shifts of the tissue, which correspond to (approximately) linear phase errors in k-space of the stimulated echo.8 The third artifact type comes from an image-domain phase shift (not a position shift) of the stimulated echo due to breathing (i.e., the breathing-induced tissue displacement that is encoded into the phase of the stimulated echo). This image domain phase shift corresponds to a constant phase error in k-space of the stimulated-echo signal and manifests as signal loss (as demonstrated later in this paper). The first and third types are specific to stimulated-echo imaging. The second type is common among most MRI methods and is predicted and accounted for using conventional models that describe the effects of breathing in MRI.12
Prior work developing self-navigated DENSE introduced the match-making method,8 which effectively deals with suppression of the T1 echo in free-breathing phase-cycled DENSE and uses stimulated echo–based image navigators (ste-iNAVs) to estimate and correct for in-plane position shifts due to breathing (i.e., the first and second types of artifacts, respectively, as discussed previously). However, the encoding of respiratory motion into the phase of the stimulated echo (the third type of artifact) has yet to be well described and corrected.13
Previous studies have demonstrated the use of deep learning for motion estimation and integration of motion fields in a motion-compensated reconstruction.14, 15 In this paper we (1) present a new model that fully describes the effect of breathing (approximated as rigid translation) in cine DENSE and (2) develop a deep-learning method for the correction of respiratory-induced constant phase shifts that can be combined with match-making and ste-iNAVs to enable accurate free-breathing self-navigated cine DENSE.
2 THEORY

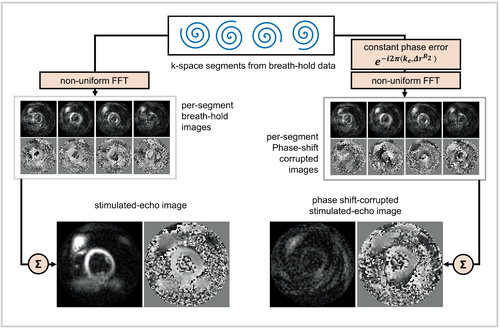
Because the constant phase errors originate from both in-plane and through-plane motion in DENSE, where it is common to use through-plane gradients to help suppress the T1 echo,11 2D ste-iNAVs are ineffective for estimating these errors. For this reason, we investigated a deep learning solution to correct for these artifacts.
3 METHODS
To validate the new motion model, we scanned a non-deformable phantom with rigid motion and compared the images with theoretical predictions. We also performed simulations to computationally validate the new motion model on DENSE images that included both respiratory and cardiac motion. To develop and evaluate the deep network for artifact correction, we acquired breath-hold DENSE data sets and, making use of the validated motion model, used computer simulations to generate training data consisting of ground-truth DENSE data (breath-hold data) and paired DENSE images with simulated motion artifacts. Next, a deep network (DENSE-RESP-NET) was trained to perform motion correction and was evaluated in healthy subjects and heart disease patients.
3.1 Phantom experiments
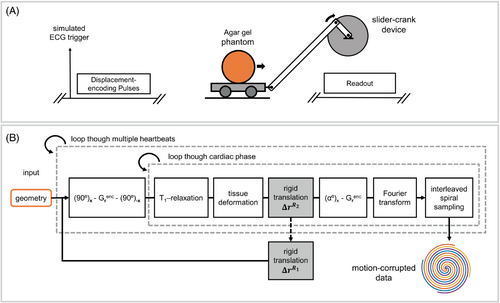
We performed phantom experiments to validate the DENSE motion model provided by Equations (2-4). The diagram in Figure 3A shows the experimental setup used for the phantom experiment. An agar gel–filled spherical phantom was positioned on a wagon that was moveable inside the bore of the magnet using a slider-crank device. Because in-plane motion before the application of the displacement-encoding pulses, , causes linear phase errors that can be corrected using ste-iNAVs, we validated the model for motion that occurs during the time between the application of the initial displacement-encoding pulses and the readout module, . Imaging was performed on a 3T MRI system (Magnetom Prisma; Siemens Healthineers) with an 18-channel phased-array body coil using a spiral cine DENSE sequence.16 DENSE images were acquired from a coronal cross-section of the phantom with diameter R = 16.4 mm using a previously described protocol3 and FOV = 350 × 350 mm2. Four spiral interleaves per image were acquired corresponding to full sampling of k-space, where two spiral interleaves were acquired during a single heartbeat and the remaining interleaves were acquired during the following heartbeat. A simulated RR interval of 3000 ms was used. The use of longer than typical RR interval provided sufficient time to move the phantom to the exact designated positions between applications of the displacement encoding and readout pulses.
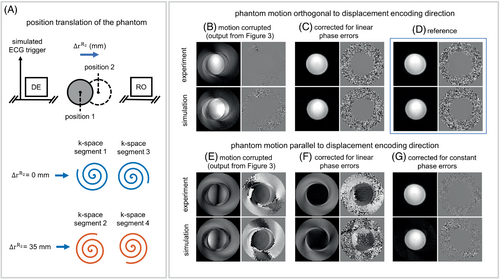
Two sets of data were acquired. The first data set was for reference, where the phantom was kept still during the acquisition. For the second data set, the phantom was moved using the slider-crank device by = 35 mm or by = 0 mm during the time between application of the displacement-encoding pulses and the corresponding readout, as illustrated in Figure 4A. To demonstrate the relationships between the direction of phantom motion and the displacement-encoding direction, a 3-point displacement encoding17 method was used to acquire three displacement measurements for each set of data. In the first and second measurements, the phantom motion was parallel to or orthogonal to the displacement-encoding direction, respectively. The third acquisition, without displacement-encoding gradients, was used to correct for the nonzero background phase. Phase-cycled data sets at matched phantom locations were also acquired to suppress the T1 echo.
For image reconstruction, phase-cycling subtraction was performed to suppress the T1 echo. Next, to simulate stimulated-echo imaging during breathing, images were reconstructed by selecting two k-space segments from the phantom position with = 0 and combining them with two segments from the data set with = 35 mm. Using the displacement-encoding frequency, ke = 0.10 cycle/mm, these parameters induce a phase shift (0.1 cycle/mm × 35 mm = 3.5 cycles) corresponding a constant phase error of for the second set of k-space segments. The motion-corrupted k-space data were corrected for linear phase errors and constant phase errors using Equation (3). After motion correction was performed in k-space, images were reconstructed using the nonuniform fast Fourier transform.18
3.2 Bloch-equation DENSE simulations incorporating cardiac and respiratory motion
We also performed simulations to computationally investigate the effects of respiratory motion on DENSE images in combination with simulated cardiac deformation (which was not feasible in our non-deforming phantom). Figure 3B shows a diagram of the DENSE Bloch equation simulations incorporating cardiac and respiratory motion.16 The simulator received the geometry of a computational phantom and its corresponding temporal deformation function19 as input. For each voxel in the phantom, the Bloch equations were solved to compute the magnetization with respect to time, accounting for the application of RF and gradient pulses in the displacement-encoding and readout modules, as well as for the effects of motion and T1 relaxation. The 2D Fourier transform was applied to the transverse magnetization to compute its k-space representation. The simulated k-space signal was sampled using interleaved spiral trajectories computed according to the parameters used to acquire data in the phantom experiment.
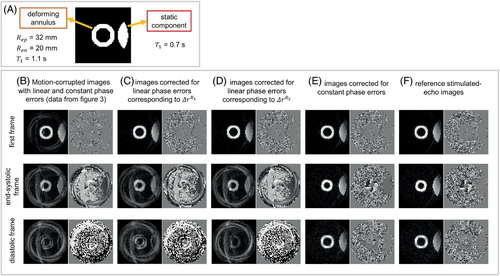
The T1 of the deforming annulus and the static component were set to 1.1 and 0.7 s, respectively. The parameters of the simulation were selected to mimic the following imaging parameters: FOV = 200 × 200 mm2, spiral readout length = 5.6 ms, in-plane spatial resolution = 3.4 × 3.4 mm2, and variable flip angle with final α = 15°. Balanced three-point displacement encoding16 was used for 2D in-plane displacement measurements, and four spiral interleaves per image with two interleaves acquired per cardiac cycle were used. The simulated cardiac cycle had a duration of 0.8 s. Match-making phase-cycled acquisitions were also simulated for suppression of the T1 echo using matched respiratory positions for the phase-cycling pairs. The displacement-encoding frequency was set to 0.06 cycles/mm.
The simulated motion-corrupted data were corrected for linear and constant phase errors (following Equations [3] and [4]) using the known motion, and the resulting k-space data were reconstructed using the nonuniform fast Fourier transform.18 The motion-corrupted and motion-compensated images were compared with reference images without motion.
3.3 Acquisition of training data for the convolutional neural network
In addition, we measured the magnitude and period of superior–inferior motion of the diaphragm in the human subjects using 15-s navigator scout scans to provide an estimate of the magnitude and variation of respiratory motion. The average and range of the superior–inferior motion was measured as 15.6 mm and 14 mm, respectively, for magnitude (peak-to-peak) and 3.83 s and 3.28 s, respectively, for the period.
3.4 Architecture of the deep learning model
We used an instance of U-Net22, 23 as the backbone of the deep learning model. Previous work has shown the advantage of exploiting temporal dependencies of MR cine images into deep learning models using either temporal convolutions or recurrent connections.24-26 Because the long/short-term memory27, 28 model as a special case of recurrent neural networks has proven stable and powerful for modeling long-range dependencies,29 the convolutional modules in the generic U-Net were replaced with convolutional long/short-term memory models to exploit the time correlations of the cine DENSE data. More details are provided in the Supporting Information.
3.5 Training of the constant-phase correction deep learning model (DENSE-RESP-NET)
We postulated that the artifacts caused by the respiratory motion–induced constant phase error could be corrected using a deep learning model. Referring to Figure 6, to train the model, phase error–corrupted DENSE images, , were generated using Equation (4) and the following steps: (a) Respiratory motions were simulated using the sine wave defined in Equation (5); (b) the corresponding phase errors were computed using the simulated motion, , where i denotes the ith k-space segment; and (c) the phase error terms were applied to breath-hold DENSE data according to Equation (4). The respiratory-induced translations, , were calculated using , where denotes the position of the heart when the nth frame is acquired, and denotes the position of the heart at the beginning of the corresponding heartbeat. The resulting motion-corrupted and the corresponding uncorrupted breath-hold images were used to train the model serving as input and ground truth, respectively. A diagram of the training is shown in Figure 6. The breath-hold data from 23 healthy volunteers and 4 heart-disease patients with three slices per subject and three displacement measurements (x, y, and background) per slice provided 243 multiphase cine DENSE images for training. To accommodate for unseen geometries, spatial translations and rotations in addition to image flipping were used to augment the training set, which provided 972 cine images after data augmentation. For each set of cine images, various breathing patterns were simulated, in which the amplitude and the period of the sine wave were randomly selected from uniform distributions mm and times/min, respectively. Thirty and six combinations of sine wave amplitude and frequency, respectively, were simulated per set of cine images, resulting in 29 160 and 5832 pairs of cine images for training and validating the model, respectively. The variation of amplitude and period from one heartbeat to another provided additional variations in the simulated respiratory patterns. The real and imaginary parts of images were separated and formatted as two-channel data. Training was performed on a Nvidia Tesla V100 GPU core for 72 h. The training was formulated as minimization of the mean squared error using the ADAM optimizer. We used a batch size and learning rate of 1 and 0.001, respectively. The trained model is referred to as DENSE-RESP-NET.
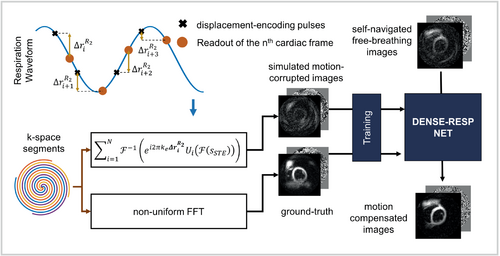
3.6 Testing of the methods using prospectively acquired free-breathing DENSE
To test the trained model, we acquired free-breathing DENSE images from 9 healthy volunteers (age = 25.9 ± 3.7; 44% female) and 3 heart-disease patients (age = 60.5 ± 10.8; 100% male). Free-breathing data were acquired using a modified spiral DENSE sequence8 as previously described.8, 30 A temporal diagram of data acquisition is provided in the Supporting Information (Figure S3). To facilitate T1-echo suppression using match-making, the acquisition of each interleaf was repeated 4 times using an average loop (number of averages = 4 in Equation [6]), resulting in a 50-heartbeat imaging protocol while keeping all other parameters the same as those used for breath-hold data. Free-breathing and breath-hold short-axis images at basal, midventricular, and apical levels were acquired for each subject.
The free-breathing data were reconstructed and processed by the trained model using the following operations. First, for each time frame, phase-cycled spiral interleaves at matched respiratory positions were identified using the match-making algorithm8 and were subtracted to suppress the T1 echo. Second, ste-iNAVS were used to correct for intersegment in-plane position shifts and corresponding linear phase errors, as previously described.8 Third, the resulting spiral multicoil cine DENSE data were reconstructed as described previously.8, 23 These images are referred to as self-navigated free-breathing images. Fourth, the self-navigated free-breathing images were processed by DENSE-RESP-NET.
3.7 Evaluation of DENSE-RESP-NET
DENSE-RESP-NET was evaluated on volunteer and patient data by comparing self-navigated free-breathing images processed by DENSE-RESP-NET to unprocessed self-navigated free-breathing images, free-breathing data with multiple averages without any type of self-navigation or phase correction, and to breath-hold data, which served as the reference standard.
In addition, the DENSE-RESP-NET processed, self-navigated free-breathing, and breath-hold images were analyzed for strain using established methods.32-34 The averaged free-breathing images did not undergo strain analysis, because the image quality was so poor that the semi-automated myocardial segmentation procedure generally failed for these data sets. Because end-systolic circumferential strain is the most commonly reported metric for cardiac strain MRI, we performed Bland–Altman analysis for this parameter. We also analyzed the early diastolic circumferential strain rate, as the constant phase error has a greater impact on diastolic frames.
4 RESULTS
4.1 Validation of the DENSE signal model using a moving, non-deformable phantom
Motion-corrupted DENSE magnitude and phase images from experimental and simulation studies are shown in Figure 4B–G for the cases in which the moving, non-deforming phantom motion was parallel (Figure 4E–G) to or orthogonal (Figure 4B,C) to the displacement-encoding direction. When motion is orthogonal to the displacement-encoding direction, only linear phase errors and blurring/ghosting artifacts occur, and the artifacts are eliminated by correcting the per-segment linear phase errors. When the motion is parallel to the displacement-encoding direction, the motion is encoded into the phase of the stimulated echo and causes constant phase errors in k-space in addition to the linear phase errors. Motion-corrupted images corrected for just the known linear phase errors are shown in Figure 4F, and the remaining artifacts in the images are due to the constant phase errors. The images corrected for the remaining constant phase errors, using Equation (4), are shown in Figure 4G.
4.2 Simulated respiratory motion–induced artifacts in DENSE images of a deforming digital phantom and their correction using the proposed model
Figure 5B–F shows DENSE magnitude and phase images from the deforming computational digital phantom generated using Bloch equation simulations. Simulated motion-corrupted images are shown at the initial, end-systolic, and middiastolic phases of the cardiac cycle. Using Equations (3) and (4), linear phase corrections corresponding to and and constant phase corrections were applied to the simulated motion-corrupted data, and the resulting images are shown in Figure 5C–E. The reference images are shown in Figure 5F. The simulated respiratory motion led to signal loss in the magnitude images and phase corruption in the phase images. However, using the known respiratory motion and the per-segment linear and constant phase corrections according to Equations (3) and (4), the displacement-encoded stimulated-echo images are recovered from their respiratory motion–corrupted versions, and the effects of cardiac motion on the image phase are preserved.
4.3 The deep learning model can correct the respiratory motion–induced artifacts in self-navigated free-breathing DENSE
Example diastolic magnitude and phase images from self-navigated free-breathing DENSE processed with DENSE-RESP-NET are shown in Figure 7 for a healthy subject (Figure 7I,J) and a heart-disease patient (Figure 7K,L), and for comparison the same images are shown for averaged free-breathing (Figure 7A–D), self-navigated free-breathing without DENSE-RESP-NET (Figure 7E–H), and breath-hold DENSE (Figure 7M–P). Signal loss in the magnitude images (Figure 7E,G) and phase corruption in the phase images (Figure 7F,H) of the self-navigated free-breathing data due to respiratory motion are readily apparent in the diastolic images in which breathing has a larger effect. The deep learning model restored the signal loss and corrected the phase values in the DENSE-RESP-NET-processed magnitude (Figure 7I,K) and phase (Figure 7J,L) images, respectively.
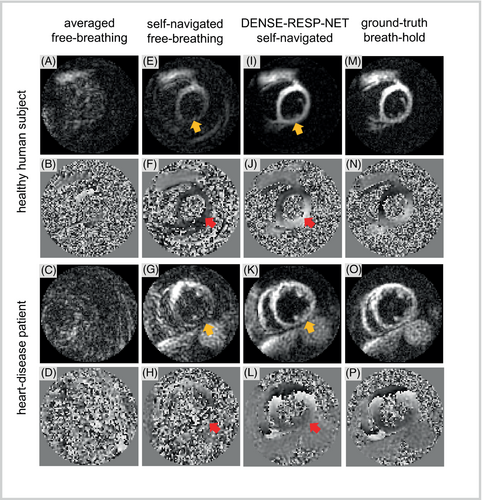
The bar plot in Figure 8 summarizes the comparisons of the phase SNR. The phase SNR assessed for DENSE-RESP-NET-processed images was significantly higher compared with the self-navigated and averaged free-breathing images and was comparable to those of breath-hold images. In addition, the self-navigated free-breathing images had significantly higher phase SNR than the averaged free-breathing data. The qualitative assessment scores (mean ± SD) were 4.42 ± 0.58, 4.23 ± 0.42, 2.88 ± 0.76, and 1.74 ± 0.80 for breath-hold, DENSE-RESP-NET-processed, self-navigated free-breathing, and averaged free-breathing phase and magnitude images, respectively (p < 0.001). The post-hoc Mann–Whitney U test showed significant differences among all four groups except breath-hold and DENSE-RESP-Net-processed images.
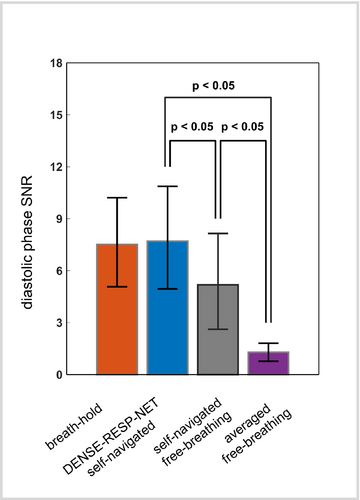
Figure 9 shows segmental circumferential strain-time curves and global circumferential strain-rate time curves for the breath-hold, the DENSE-RESP-NET-processed, the self-navigated free-breathing, and the averaged free-breathing images computed from the example images shown in Figure 7. Overall, the DENSE-RESP-NET processed curves are in close agreement with breath-hold curves for both segmental strain and the global strain rate. The strain and strain rate curves from the self-navigated free-breathing images are in close agreement with those of the reference and the DENSE-RESP-NET processed images early in the cardiac cycle. However, the agreement worsens in diastole. The strain and strain-rate curves from the averaged free-breathing images were unreliable compared with those of reference images.
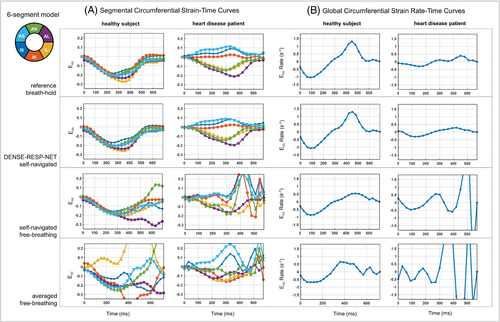
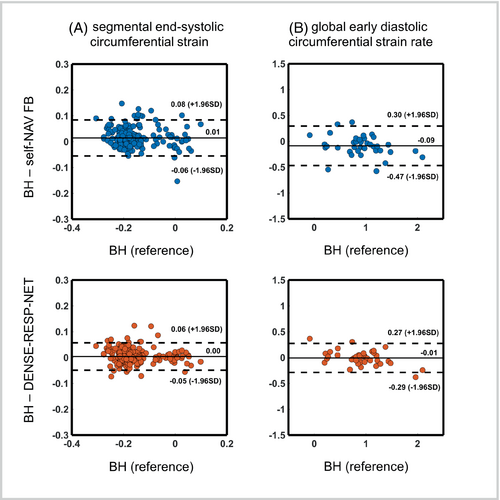
Figure 10 summarizes the Bland–Altman analyses for the segmental end-systolic circumferential strain and the global early diastolic circumferential strain rate for the 36 DENSE slices acquired from the 9 volunteers and 3 heart-disease patients. The end-systolic segmental strain shows modestly better agreement for DENSE-RESP-NET compared with unprocessed, self-navigated, free-breathing images with respect to the breath-hold images. For early diastolic strain rate, agreement and accuracy are substantially improved in favor of the DENSE-RESP-NET-processed images.
5 DISCUSSION
The main contributions of this study are that (a) a new model was introduced and validated that describes the effects of rigid translational motion due to breathing on the magnitude and phase of the DENSE signal, and (b) the new motion model was used to train a deep convolutional neural network, DENSE-RESP-NET, to correct the constant phase artifacts in free-breathing cine DENSE attributed to the encoding of respiratory motion into the phase of the stimulated echo. When used in concert with the match-making method for suppression of the T1 echo and ste-iNAVs for correction of linear phase errors in k-space related to object motion, DENSE-RESP-NET-corrected self-navigated free-breathing DENSE images show good agreement with breath-hold DENSE images, and the corresponding strain data show good agreement with breath-hold strain data for the evaluation of both systolic and diastolic parameters.
Our phantom experiments and Bloch equation simulations using nondeforming and deforming phantoms were used to validate the DENSE motion model of Equations (2-4). For the phantom and computational experiments, the object motion was known, and our results in Figures 4 and 5 showed that correcting the phase of the k-space data according to Equations (2-4) successfully removed the signal loss and phase-corruption artifacts from the images. Together, these results demonstrate the validity of Equations (2-4) for describing the effects of motion on segmented stimulated-echo images.
In the phantom experiments, we made measurements corresponding to motion parallel to and orthogonal to the direction of the displacement-encoding gradients, but we did not explore motion in the through-plane direction. As indicated by Equations (2-4), the nature of the artifact (being due to a constant phase shift) is consistent, regardless of whether the motion occurs in the in-plane or through-plane direction. Thus, the experimental results shown in Figure 4, investigating phantom motion orthogonal to and parallel to the displacement-encoding direction, are sufficient for demonstrating the effects of on DENSE magnitude and phase images.
The present study used the new DENSE motion model in combination with breath-hold DENSE k-space data to generate synthetic data that were used to train DENSE-RESP-NET. To our knowledge, other than using synthetic data as presented in this study, there is no other time-efficient and cost-efficient way to obtain training data for this task. Therefore, for this task it was essential to understand the physics underlying the artifacts, as a description of the underlying physics was needed to generate training data and facilitate a deep learning solution. Other studies have also demonstrated the use of simulated respiratory motion artifacts for training deep learning models for other MRI methods.35-37 Also, the amount of training data and data augmentation used in this study to train the DENSE-RESP-NET was determined to assure stability of training and validation curves, avoid overfitting, and reduce the training time. We did not observe a significant difference in performance when more training data were used.
Because DENSE generally uses in-plane displacement-encoding gradients as well as through-plane dephasing gradients, breathing motion in any direction will manifest as the signal loss and phase corruption artifacts described in Equations (2-4). Because the DENSE-RESP-NET was designed to compensate for such artifacts without knowledge of the specific magnitude or direction of the respiratory motion, the model corrects for both in-plane and through-plane breathing motion.
A limitation of the current approach was that the respiratory motion was considered as rigid translation. Although this first-order approximation significantly compensated for the respiratory-induced artifacts, future work could consider nonrigid tissue deformation and potentially improve the results. Another limitation was that long-axis images were not included in the training and testing data. Finally, the in vivo data used in this study were acquired with a specific set of acquisition parameters. While we showed good performance of free-breathing cine DENSE with match-making, ste-iNAVs and DENSE-RESP-NET using a specific protocol, a generalized deployment of the deep learning model on self-navigated free-breathing images with different acquisition parameters and various types of heart disease may require retraining of the model. Future studies may evaluate the current model in a larger number of heart-disease patients and evaluate its generalizability.
6 CONCLUSIONS
A new model was introduced to describe the effects of breathing on the magnitude and phase of the DENSE signal, and it was used to train DENSE-RESP-NET to correct for breathing-associated constant phase errors. When used in combination with the match-making method for suppression of the T1 echo and ste-iNAVs for correction of linear phase errors, DENSE-RESP-NET-corrected, self-navigated, free-breathing DENSE images and myocardial strain data show good agreement with breath-hold DENSE.
CONFLICT OF INTEREST
Mohamad Abdi is an employee of Siemens Healthineers (Malvern, PA, USA).




