Resveratrol and prostate cancer: Promising role for microRNAs
Abstract
Scope: Resveratrol (Res) has anticancer activity in prostate cancer (PCa), which can be attributed to modulation of microRNAs (miRNAs/miRs). miRNAs/miRs are small non-coding RNAs that negatively regulate gene expression. We have analyzed differential miRNA expression in PCa cells treated with Res.
Methods and results: Using miRNA microarrays we found that 23 miRNAs were significantly down-regulated and 28 miRNAs were significantly up-regulated after Res treatment. The down-regulated miRs included miR-17-92 and miR-106ab clusters with well recognized oncogenic properties while the up-regulated miRs included several tumor suppressors. Selected miRs were verified by qRT-PCR, including miR-17, miR-20a, miR-20b, miR-106a and miR106b. Since these miRNAs target PTEN (phosphatase and tensin homolog deleted on chromosome 10), we performed Western blot to confirm up-regulation of PTEN in PCa cells. In addition, using TargetScan database, we have identified putative mRNA targets for Res-induced down- and up-regulated miRs. Using a bioinformatics approach, we generated gene networks specifically altered by Res-regulated miRNAs.
Conclusion: Our results indicate that the dietary compound Res may play an important role in prostate carcinogenesis through modulation of miRNA expression: Res down-regulated oncogenic miRs and up-regulated tumor suppressor miRs in PCa cells. Further in-depth studies are necessary in order to fully recognize the beneficial miRNA-mediated effects of Res in PCa.
1 Introduction
Prostate cancer (PCa) is the most common male malignancy and the second cause of cancer-related death in the US. Nutritional chemoprevention is a particularly promising approach for PCa because of its slow progression and predominance in elderly men. Resveratrol (Res) has been proposed as an especially suitable agent for PCa due to its specific androgen receptor (AR)-related effects 1. In addition, epidemiological data indicate a reduced PCa risk associated with red wine consumption, attributable mostly to high Res content 2.
Res (3, 5, 4'trihydroxy-trans-stilbene), a natural phytoalexin, is known for its cardioprotective, anti-inflammatory and anticancer activities 3. Res has pleiotropic anticancer activities that include induction of cell cycle arrest and apoptosis 4-6 and inhibition of angiogenesis, invasion and metastasis 7-10. Little is known on the epigenetic mechanisms of Res chemoprevention and anticancer effects. Accumulated data suggest that beneficial effects of dietary polyphenols may be in part attributed to their epigenetic properties, including changes in DNA methylation, regulation of histone modifications and changes in the expression of microRNAs (miRNAs/miRs) 11. However, this is an area that is mostly underexplored, particularly in PCa 12. Currently, epigenetic mechanisms of Res are associated with the ability of Res to activate histone acetyl transferase (HAT) p300 and histone deacetylase sirtuin 1 (SIRT1) 13, 14. The exact mechanism by which Res activates SIRT1 remains controversial 15, 16. The apparent paradox that Res can simultaneously activate histone acetyl transferases and a deacetylase remains unexplained. In contrast, we recently demonstrated that Res acts as natural indirect histone deacetylase (HDAC) inhibitor by reversing the activity of co-repressor nucleosome remodeling and deacetylation complex (NuRD) involved in histone deacetylation and gene-specific transcriptional regulation: Res down-regulates metastasis-associated antigen 1 (MTA1), a protein that has pro-survival, anti-apoptotic, invasive and pro-angiogenic properties in PCa 17, 18. We further demonstrated that MTA1 silencing through RNA interference significantly sensitized the PCa cells to Res-dependent p53 acetylation and apoptosis 19.
Another important epigenetic network of gene regulation is represented by miRNAs 11, 20. Recently, few reports indicated that dietary factors play an important role in the process of carcinogenesis through modulation of miRNA expression 21, 22. Therefore, modulation of miRNA expression may contribute to the cancer-protective effects of Res known as a potential chemopreventive and therapeutic agent in PCa. miRNAs are small (∼18–22 nucleotides), non-coding, single-stranded, sequence-specific RNAs that negatively regulate gene expression by binding complementary sequences in the 3′ untranslated regions (UTRs) of target mRNA and inhibiting stability and/or translation of mRNA 23, 24. miRNAs act as oncogenes or tumor suppressors in cancer 25. Oncogenic miRNAs (oncomiRs) that are overexpressed in different cancers usually promote tumor development by inhibiting tumor suppressor genes and genes that control cell differentiation and apoptosis. In contrast, tumor suppressor miRNAs have decreased expression in cancers and usually prevent tumor development by negatively affecting oncogenes and promoting genes that control cell differentiation and apoptosis. Various miRNA expression profiling studies reported on both up- and down-regulation of miRNAs in PCa tissues compared to normal prostate tissue or benign peripheral zone tissue from radical prostatectomy specimens 26, 28. Some changes in miRNAs have been correlated with Gleason score and clinical recurrence 29, 30. Thus, differential miRNAs in PCa can be useful diagnostic and prognostic indicators. Because of high enthusiasm about the use of Res in human chemoprevention 31, 32, the ability of Res to alter miRNAs offers new perspectives for Res chemoprevention and anticancer activities. However, there are no reports on Res-dependent alterations of miRNA expression in PCa.
To our knowledge, this is the first study to look at changes in miRNA levels by Res in PCa. In this study, we showed that Res significantly down-regulated several miRNA oncogenic clusters, up-regulated some known tumor suppressors and modulated other miRNAs whose biological role has yet to be established.
2 Materials and methods
2.1 Materials
Res was purchased from Sigma (St. Louis, MO, USA). Stock solution of Res was made using 100% ethanol and kept at 4°C, in the dark. Lymph node cancer of prostate (LNCaP) and Du145 cells were maintained in RPMI medium containing 10% FCS as described previously 18, 19. Cells were maintained in phenol red-free media with 5% charcoal-stripped calf serum overnight prior to Res treatment.
2.2 Array-based miRNA expression profiling and analysis
LNCaP cells were maintained in RPMI 1640 medium containing 10% FCS as we previously described 18, 19. When cells achieved about 60% confluence, the media was replaced with phenol red-free RPMI supplemented with 5% charcoal-stripped calf serum overnight to eliminated hormonal background that might interfere with Res effects. Res or EtOH-vehicle was added to the media for the next 24 h after which RNA was isolated using miRNeasy Mini Kit (Qiagen, Valencia, CA). We used miRNA microarray detection service from LC Sciences (Houston, TX, USA) using MRA 1001 chip, which contains 894 human miRNA probes listed in Sanger miRBase Release 14.0 (http://www.sanger.ac.uk/Software/Rfam/mirna/). RNA integrity and purity was checked before analysis. The miRNA-enriched samples were then labeled with fluorescent dyes for two color detection (Cy3 and Cy5). Hybridization, washing and detection of miRNA array chips were controlled to optimize signal. After background subtraction and normalization, data analysis was performed, and a list of up- and down- regulated transcripts with statistical significance of the p-values <0.01 was provided.
2.3 Identification of miRNA putative target genes and network analysis
TargetScan database, version 5.1 (www.targetscan.org) was used for identification of putative targets for identified miRNAs. This algorithm predicts miRNA targets on the basis of sequence complementarity and thermostability 33. The overall target specificity is expressed by a context score 34 that combines the complementarity of seed sequence, site position and local adenine/uridine (AU) content to evaluate site efficacy between miRNAs and mRNA targets. To enhance the accuracy for miRNA target prediction and select high confidence targets, only genes with total context score ≤−0.5 were included in our analysis. We generated regulatory networks and visualization using Bayesian modeling for the putative targets of two co-regulatory miRNAs sets: oncogenic and tumor suppressor. We then integrated these genes onto a global molecular network (gene ontology (GO) database), which identifies the biological functions that are most significant to the genes in the network.
2.4 Validation of miRNA expression by real-time RT-PCR
RNA was isolated using miRNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. qRT-PCR was performed using mercury LNA™ Universal RT miRNA PCR (Exiqon, Woburn, MA). After first-strand cDNA synthesis, the real-time amplification was carried out using cDNA template (8 μL), SYBR Green master mix (10 μL) and miRNA specific primer mix (2 μL) in a final volume of 20 μL. The condition for PCR was 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 1 min, performed in a CFX96 Real-Time System (Bio-Rad). All primers used for qRT-PCR were purchased from Exiqon. SNORD44 was used as an internal reference standard. Fold change of normalized miRNA was calculated using the 2−ΔΔCt for each miRNA by double-delta Ct method 35. Student's two-tailed paired t-test was performed to analyze results from qRT-PCR.
2.5 Western blot
Western blot analysis was performed as described previously 18, 19, 36. Briefly, the cells were maintained in regular RPMI 1640 media, and then media was changed to phenol red-free supplemented with stripped serum 24 h before treatment with Res. Cells were harvested using lysis buffer. Equal amounts of protein were size-fractionated by 7.5% Tris-HCl Ready gels (Bio-Rad Labs, Hercules, CA) and transferred to polyvinylidene fluoride (PVDF) membrane. The membranes were then blocked with Tris buffered saline (TBS)-Tween and 5% dry milk and subsequently probed with anti-PTEN (PTEN, phosphatase and tensin homolog deleted on chromosome 10) and anti-β-actin antibodies (Santa Cruz, Santa Cruz, CA). Signals were visualized using enhanced chemiluminescence. Images were quantified using Image J software.
3 Results and discussion
3.1 Res alters miRNA expression in human PCa cells
Res induces apoptosis, inhibits cancer cell growth, angiogenesis and metastatic potential of PCa cells in vitro and in vivo 19, 37-40. These effects of Res could be mediated via the regulation of miRNAs. As mentioned, studies reported on both up- and down-regulation of miRNAs in PCa 26-28. Moreover, the link between miRNAs changes and the development and progression of PCa has been established 41-43. Since Res is an anticancer agent in PCa, Res-induced down-regulated miRNAs must function as oncomiRs whereas Res-induced up-regulated miRNAs must function as tumor suppressors. We performed miRNA microarray analysis of Res-treated LNCaP cells and found that 51 miRNAs were modulated, from which 23 miRs were significantly down-regulated (oncomiRs) and 28 miRs were significantly up-regulated (tumor suppressor miRNAs) following treatment with 50 μM Res (Fig. 1). Each group of statistically significant miRNAs is represented as two subgroups: above the horizontal line are transcripts with high signal intensity and below are transcripts with low signal intensity (Fig. 1B). We are interested in the differences between expression levels for a given miRNA in untreated and Res-treated cells independent of its basal levels of expression. In fact, for most miRNAs with low signals (low abundance), the changes in expression after Res treatment were more dramatic than for miRNAs with higher abundance. These changes could be of particular interest and should not be overlooked.
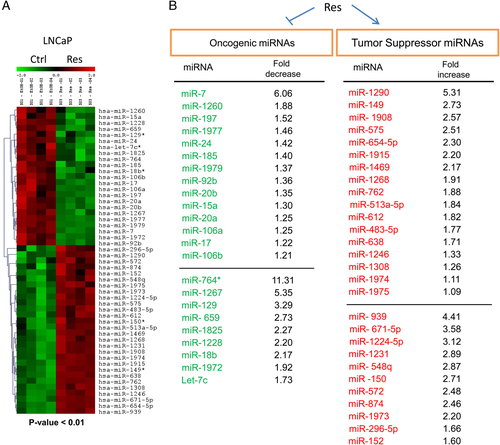
Significant changes in miRNA expression in Res-treated LNCaP cells. (A) Heat map showing differential expression of miRNAs (hsa-miRs) in LNCaP treated with either EtOH (control, trl) or 50 μM Res. p<0.01. The relative transcript abundance of each miR is color coded: green indicates low expression; black indicates intermediate and red indicates high expression. (B) Results of analysis ranked by fold change. A total of 23 miRs (oncomiRs) were significantly down-regulated (green) and 28 miRs (tumor suppressor miRNAs) were significantly up-regulated (red). Fold decrease/increase was calculated from log2 Res/Ctrl values. Horizontal lines in each set separate two subgroups of miRNAs: transcripts with higher abundance (above the line) and transcripts with low abundance (below the line). Note: hsa-miR hereafter referred to as miR.
We performed a vigorous literature and web search to draw a parallel between our findings and already known roles of miRNAs in human malignancies, particularly in PCa. First, we generated two lists of potential gene targets for identified onco- and tumor suppressor miRNAs using TargetScan database (http://www.targetscan.org), which predicts target mRNAs by seed sequence complementarity (Supporting Information data, ST1 and ST2). To select high confidence targets we used single criterion of the context score threshold and filtered out targets with score >−0.5. The total number of putative targets predicted for Res-regulated oncogenic and tumor suppressor miRNAs are shown in Table 1. We then compared our data to the miRNA expression reported by others, in which miRNAs were classified as oncogenes or tumor suppressors. We first searched for oncomiRs. Remarkably, Res significantly down-regulated five (miR-17, miR-18b, miR-20a, miR-20b and miR-92b) from six members of the c-MYC-activated oncomiR-17-92 cluster 44. This cluster is amplified in lung cancer and lymphoma, frequently overexpressed in various cancers, including PCa, and known to accelerate angiogenesis and promote tumor formation in mouse models 25, 26, 45-48. As an example, overexpression of miR-20a inhibited apoptosis in PCa cell lines and inhibition of miR-20a resulted in increased apoptosis after doxorubicin treatment 49. Overexpression of cell cycle transcription factor E2F1, which is one of the targets of the miR-17-92 cluster, induced apoptosis in LNCaP cells 47, 50. Further, Res inhibited miR-106a and miR-106b oncomiRs that are reported to be highly expressed in PCa and mediate antiapoptotic activity by targeting p21/WAF1 and E2F1 41. Notably, both miR-17-92 and miR-106b-25 clusters are regulators of a critical tumor suppressor gene PTEN, and miRs from these clusters directly associate with PCa tumorigenesis 51. For example, the miR-106b-25 locus in chromosome 7 is overexpressed and amplified in PCa. Moreover, miR-106a-25 expression is inversely correlated with PTEN abundance in human PCa tissues 51. Since PTEN is frequently defective in PCa as its deletions/mutations are found in 30% of primary and in 63% of metastatic disease 52, 53, Res rescue of PTEN by down-regulation of the oncogenic PTEN-targeted miRNA-network is of considerable interest.
| Oncogenic miRNAs | Targets (n)a) | Tumor suppressor miRNAs | Targets (n)a) |
|---|---|---|---|
| miR-7 | 33 | miR-1290 | 46 |
| miR-1260 | 21 | miR-149 | 17 |
| miR-197 | 10 | miR-1908 | 0 |
| miR-1977 | 0 | miR-575 | 4 |
| miR-24 | 36 | miR-654-5p | 3 |
| miR-185 | 34 | miR-1915 | 0 |
| miR-1979 | 0 | miR-1469 | 0 |
| miR-92b | 0 | miR-1268 | 1 |
| miR-20b | 3 | miR-762 | 0 |
| miR-15a | 10 | miR-513a-5p | 47 |
| miR-20a | 21 | miR-612 | 5 |
| miR-106a | 0 | miR-483-5p | 0 |
| miR-17 | 68 | miR-638 | 0 |
| miR-106b | 3 | miR-1246 | 14 |
| miR-764 | 0 | miR-1308 | 4 |
| miR-1267 | 8 | miR-1974 | 0 |
| miR-129 | 0 | miR-1975 | 0 |
| miR-659 | 49 | miR-939 | 3 |
| miR-1825 | 6 | miR-671-5p | 10 |
| miR-1228 | 4 | miR-1224-5p | 7 |
| miR-18b | 1 | miR-1231 | 3 |
| miR-1972 | 0 | miR-548q | 0 |
| let-7c | 1 | miR-150 | 17 |
| miR-572 | 0 | ||
| miR-874 | 3 | ||
| miR-1973 | 0 | ||
| miR-296-5p | 4 | ||
| miR-152 | 5 |
- a) a) Target numbers are based on total context score ≤−0.50. Table is created using TargetScan database (www.targetscan.org).
Among the most significantly down-regulated miRNAs by Res were miR-7 (6.06-fold decrease) and miRs from a second subgroup of oncomiRs with a fold decrease ranging from 1.92 to 11.31 (Fig. 1B). The changes of low abundant miRNAs after Res treatment were more pronounced than in miRNAs with high signals and might have more biological significance. However, little is known about the cancer-related functions of most of the identified miRs in this subgroup. High expression of miR-1228 was reported in malignant mesothelioma compared to normal samples 54. Expression of miR-129 (and corresponding low SRY-related HMG box (SOX) 4 levels) in bladder cancer was found to be associated with poor outcome 55. On the other hand, the expression of miR-129-2 was lost in primary endometrial tumors, and reactivation of miR-129-2 in cancer cells resulted in decreased SOX4 and reduced proliferation 56. The biological functions of miR-764, miR-1267, miR-659, miR-1825 and miR-1972 have not been characterized in cancer. However, while miR-764 and miR-1972 showed no identified putative targets (TargetScan), miR-659, miR-1825 and miR-1267 indicated numerous targets that are implicated in cancer (Table 1). For example, putative targets of miR-659, suppressor of cytokine signaling (SOCS) protein family, play an important role in PCa: SOCS1 is expressed in PCa and acts as a negative growth regulator 57 whereas androgen-regulated SOCS2 seems to have a role in hormonal progression to androgen-resistant disease 58.
Other down-regulated miRNAs by Res were miR-7, miR-197 and miR-24. The role of miR-7 in tumorigenesis is complex and undecided: it has been reported as both an oncomiR and tumor suppressor. Overexpression of miR-7 was documented in breast and lung cancer and was associated with poor outcome of diseases 59, 60. On the other hand, miR-7 has been characterized as a tumor suppressor targeting oncogenes and a number of key signaling molecules of survival pathways in several cancers 61-63. It is likely that particular interaction between a miRNA and its target genes may be influenced by microenvironment and dependent on cancer type and cell specificity. Because Res down-regulated miR-7 in our study, we believe that miR-7 plays an oncogenic role in PCa. In agreement with this notion, prostate apoptosis response protein 4 (Par-4), a tumor suppressor protein that was isolated from PCa cells undergoing apoptosis, is linked to miR-7: in Par-4-transfected colon cancer cells, miR-7 was down-regulated among other oncomiRs (e.g. miR-18a) and the down-regulation was associated with up-regulation of apoptotic voltage-dependent anion channel protein 1 (VDAC1) 64. Further studies are needed to elucidate the role of miR-7 in PCa. High levels of miR-197 were found in small-cell and non-small-cell lung carcinoma cell lines and in non-small-cell lung cancer tumor specimens, which correlated with reduced expression of target tumor suppressor Fus1 (equivalent of human TUSC2, tumor suppressor candidate 2) 65. miR-24 is up-regulated in cervical and gastric cancers 26, 66. Importantly, miR-24 has been suggested as a potential target for gene and drug therapy to treat hormone-insensitive PCa because the down-regulation of miR-24 induced apoptosis in Du145 cells via up-regulation of its target pro-apoptotic factor Fas-associated factor 1 (FAF1) 67. To our surprise, Res down-regulated let-7c, miR-15a and miR-185 miRNAs, which have been reported as tumor suppressors 68-70. No information was found on the roles of miR-1260, miR-1977, miR-1267, miR-1825. Altogether, Res widely down-regulated miRs with oncogenic properties that are overexpressed in various cancers, including PCa.
miRNAs up-regulated by Res fall into three major groups: (i) miRs that have been reported by others to be down-regulated in different cancers (miR-149, miR-152, miR-150; miR-939, miR-575, miR-483 and miR-654); (ii) miRs that are have been reported to be up-regulated (miR-572, miR-762 and miR-1246) and (iii) miRs whose aberrant expression or regulation have not been reported before and whose biological functions have not been characterized (miR-1290, miR-1908, miR-1915, miR-1469, miR-1268, miR-1231, miR-1973, miR-548q, miR-1974, miR-1975 and miR-671-5p). Some of the miRs from this group, however, have many putative target genes according to TargetScan database (Table 1). Although knowledge about alterations and functional roles of a large part of the identified up-regulated miRNAs is still missing, functional roles have been described for some of them. Remarkably, miR-654-5p (2.30-fold increase by Res) was recently found to be among miRs that target an extended AR 3′ UTR and down-regulate AR, prostate-specific antigen (PSA) and related androgen-induced proliferation in LNCaP cells 71. Our own data on Res down-regulation of AR and PSA in LNCaP cells suggest a possible link between Res-up-regulated miR-654-5p and Res inhibition of AR and cell proliferation (unpublished data). Loss of miR-149 has been reported to cause gain of function of KCNMA1 and lysyl oxidase (LOX) oncogenes in clear cell renal cell carcinoma 72. Notably, miR-149 was down-regulated in PCa, and was part of a six-member diagnostic marker set that discriminated between malignant and normal tissues in PCa patients 29. Down-regulation of miR-150 in relapsed patients was associated with the prognosis and disease progression of pediatric acute leukemia 73. Down-regulation of miR-150 and its tumor suppressor activity has been demonstrated in mouse model of B-cell lymphoma 44. Moreover, miR-150, one of the main regulatory miRNAs of immune function, was found to be significantly down-regulated in peripheral blood and leukocytes of sepsis patients compared to healthy controls. This down-regulation was associated with increased levels of anti-inflammatory cytokine IL-18 74. On the other hand, miR-150 is shown to promote gastric cell proliferation 75. Further, reduced expression of miR-152 has been documented in nasopharyngeal carcinoma 76. miR-152 was suggested as a tumor suppressor because very low levels were found in highly invasive melanoma cell line 77, and there was a reverse correlation with high levels of micropthalmia-associated transcription factor (MITF), an oncogene involved in melanoma progression 78. In addition, hypermethylation of miR-152 was found in 34–86% of primary breast cancer tissues 79. The down-regulation of miR-575, miR-483 and miR-654 was reported in meningiomas 80 and reduced expression of miR-939 was found in tumors of long-surviving neuroblastoma patients 81; however, the role of these miRNAs remains unknown. Our results show Res up-regulation of a group of miRNAs that were reported to be up-regulated in ovarian cancer tissues (miR-572) 82, squamous cell carcinoma (miR-762) 83 and breast cancer (miR-1246) 84. miR-513a-5p was up-regulated in human bronchial epithelial cells after exposure to diesel exhaust particles 85, and was reported to repress interferon-γ-induced apoptosis 86. miR-612 was recently mentioned as one of the six putative miRNAs negatively regulating tumor suppressor p73 87. Curiously, up-regulation of miR-1224 was reported in diet-induced fatty liver models in which mice were fed with alcoholic and nonalcoholic diets 88. To date, there is no information about the alteration/function of miR-1308 and miR-874 in cancer. Taken together, Res up-regulated tumor suppressor miRs that are lost or down-regulated in various cancers, including PCa.
In summary, the finding that Res down-regulates oncomiRs and up-regulates tumor suppressor miRNAs corroborates our general idea of Res as a powerful negative modulator of miRNA-associated tumorigenesis.
3.2 Validation of selected Res-regulated miRNAs by qRT-PCR and Western blot analysis
To validate our array data, we selected several miRNAs that were down- or up-regulated by Res in LNCaP cells and performed qRT-PCR (Fig. 2A). In agreement with the array data, changes by Res in each miR showed the same trend. Specifically, miR-7, miR-17, miR-18b, miR-20a, miR-92b, miR-106a and miR106b each decreased whereas miR-150 and miR-296-5p increased in Res-treated cells. The miRs from the miR-17-92 and miRs-106a and b oncogenic clusters were of particular interest because (i) of their importance for PCa tumorigenesis 26, 49, 51 and (ii) of their down-regulation by Res. Importantly, these two clusters are regulators of tumor suppressor PTEN, which is frequently defective in PCa. Since LNCaP cells are PTEN-negative 89, 90 and Du145 cells express wild-type PTEN 91 we pursued our findings with Du145 cells and checked whether Res down-regulates miR-17-92 and -106a, b clusters in these cells. We found that Res significantly down-regulated miRs associated with PTEN regulation in Du145 cells (Fig. 2A). To find out whether Res up-regulates PTEN protein in PCa cells, we performed Western blot analysis and showed that Res up-regulated PTEN by about twofold in Du145 cells (Fig. 2B). Whether this up-regulation is a direct consequence of miRNA-related alterations still has to be established. LNCaP cells were PTEN-negative as has been previously reported 89, 90.
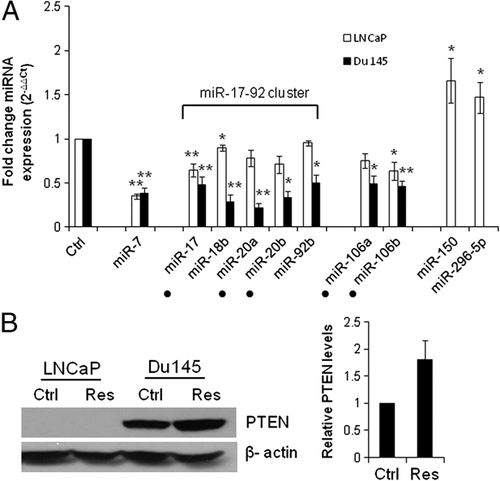
(A) Validation of selected miRNAs by qRT-PCR. LNCaP and Du145 cells were maintained in phenol red-free media/stripped serum overnight prior to treatment with 50 μM Res. RNA was isolated and qRT-PCR was performed as described in Section 2. All samples were tested in duplicate, and experiments were repeated at least four times. All samples were within the measurable range (Ct<35). ΔCt for each miR was calculated by subtracting the Ct of reference SNORD44 from the Ct of given miR. ΔΔCt was then calculated by subtracting the ΔCt of the EtOH-treated Ctrl from ΔCt of the Res-treated samples. Fold changes for the expression of miRs were calculated using the equation 2−ΔΔCt. Data are presented as the mean±SE of four or more independent experiments. *p<0.05; **p<0.01 (Student's t-test); •, indicate miRs that target PTEN according to www.microRNA.org (miRanda) (B) Res up-regulates PTEN in Du145 cells. Left, LNCaP and Du145 cells were treated with 50 μM Res and EtOH (Ctrl) when in phenol red-free media with stripped serum. PTEN expression was detected by Western blot using whole cell extracts, and levels of PTEN (55 kDa) was detected. β-actin was used as a loading control. Representative blot is shown. Right, quantification of PTEN protein levels in Du145 cells normalized to β-actin from three independent experiments: Res-treated cells express in average 1.8-fold higher PTEN than Ctrl cells. Bars, SE.
3.3 In silico network analysis of Res-regulated miRNA putative targets
It is established that multiple co-expressed miRNAs may work collectively to regulate related targets in common pathways 92. To link Res-regulated miRNA profiling data with biological consequences, we took a computational approach to examine the possible pathways collectively regulated by the co-expressed down- or up-regulated miRNAs on the basis of their predicted targets. By applying a strict criteria of ≤−0.5 context score, 308 targets were identified in down-regulated (oncogenic) set, and 193 mRNAs in up-regulated (tumor suppressor) set. miRs that did not pass the threshold of context score and showed no predicted targets (Table 1) were excluded from further analyses.
We identified a potential gene network, i.e. the interaction between potential targets for each set of Res-regulated miRNAs (Fig. 3). Within the oncomiR target group (Fig. 3A), 11 (miR-7, miR-17, miR-1228, miR-20a, miR-197, miR-15a, miR-185, miR-659, miR-1260, miR-24, miR-1825) out of 18 down-regulated miRs accounted for the 36 predicted targets that form the potential gene network, including “connectors” from the GO data base. Genes in “signal transduction”, “cell organization and biogenesis”, protein phosphorylation and metabolism form the top gene networks within this set. Within the tumor suppressor miRNA target group (Fig. 3B), seven (miR-1290, miR-654-5p, miR-1308, miR-150, miR-1231, miR-149 and miR-513a-5p) out of 16 up-regulated miRs accounted for the 11 predicted targets in the “gene transcription” network.
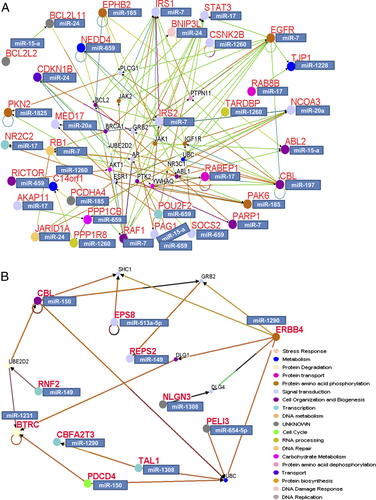
Interaction maps of miRNA potential targets (based on in silico prediction). (A) Gene network of oncomiR target group. (B) Gene network of tumor suppressor miRNA target group. Genes are represented by circles, red letters. The “big” circles indicate miR-related targets. Corresponding miRs are represented by boxes. The “small” circles indicate the intermediate genes from GO database, black letters. Lines represent interactions. Genes are color-coded according to class/function.
Our results revealed that the targets co-regulated by the Res-down-regulated oncomiRs include several key regulators of carcinogenesis such as tumor suppressor gene retinoblastoma (RB), which controls androgen signaling and PCa progression 93, tumor suppressor poly (ADP-ribose) polymerase 1 (PARP1) 94, negative growth regulators (SOCS) 57, 58 and activators of pro-apoptotic pathways (BLC2L11) 95. Interestingly, it has been shown recently that Res induces growth arrest and apoptosis in PCa cells through activation of forkhead box O (Foxo)-mediated up-regulation of BLC2L11), also known as Bim 96. It is very possible that Res up-regulation of Bim in PCa might occur in part through down-regulation of miR-24. In contrast, the targets co-regulated by the Res-up-regulated tumor suppressors include oncogenes, activators of survival pathways and inhibitors of apoptosis. For example, REPS2 (RALBP1-associated Eps domain containing 2) inhibits epidermal growth factor (EGF) signaling in advanced metastatic PCa 97 and plays a role in overcoming resistance to apoptosis in androgen-resistant PCa 98, while reactivation of v-erb erythroblastic leukemia viral oncogene homolog 4 (ERBB-4) results in apoptosis of cancer cells 99. Further studies are warranted to provide experimental validation of the complex interconnection between Res-induced co-regulated miRNAs and their targets.
4 Concluding remarks
In summary, this study provides clear evidence that Res-induced modulation on miRNA expression in PCa may contribute to the cancer-protective effects of Res. We found that Res down-regulated several important oncomiRs and up-regulated some tumor suppressor miRs in PCa cells, which potentially can lead to regulation of target mRNAs essential in carcinogenesis and cancer progression. Although the functions of most miRNAs regulated by Res have yet to be discovered, it is apparent that Res-regulated miRNA profiles provide a basis for future experimental functional studies in vitro and in vivo to identify their physiological role in PCa and to offer a novel perspective on the potential chemopreventive and chemotherapeutic benefits of Res.
Acknowledgements
We are extremely grateful to Dr. L. Kai (Northwestern University, Chicago, IL) for performing the Western blot and Ms. T. Slaughter for performing TargetScan search for target prediction. We thank Mr. S. Dias for his excellent technical assistance. We also thank Drs. L. Miele and S. Bigler for their continuous support. This study was supported in part by the Intramural Research Support Program (IRSP) of the University of Mississippi Medical Center to A.S.L.
The authors have declared no conflict of interest.




