Dispersion of synonymous codon usage patterns in hepatitis E virus genomes derived from various hosts
Xin Wang and Jing Sun contributed equally to this study.
Abstract
Hepatitis E virus (HEV) is an important zoonotic pathogen infecting a wide range of host species. It has a positive-sense, single-stranded RNA genome encoding three open reading frames (ORFs). Synonymous codon usages of viruses essentially determine their survival and adaptation to susceptible hosts. To better understand the interplay between the ever-expanding host range and synonymous codon usages of HEV, we quantified the dispersion of synonymous codon usages of HEV genomes isolated from different hosts via Vs calculation and information entropy. HEV ORFs show species-specific synonymous codon usage patterns. Ruminant-derived HEV ORFs own the most synonymous codons with stable usage patterns (Vs value <0.1) which leads to the stable overall codon usage patterns (R value being close to zero). Swine-derived HEV ORFs own more concentrated synonymous codons than those from wild boar. Compared with HEV strains isolated from other hosts, the human-derived HEV exhibits a distinct pattern at the overall codon usage (R < 0). Generally, ORF1 contains more synonymous codons with stable usage patterns (Vs < 0.1) than those of ORFs 2 and 3. Moreover, ORF3 contains more synonymous codons with varied patterns (Vs > 1.0) than ORFs 1 and 2. The host factor serving as one of the evolutionary dynamics probably influences synonymous codon usage patterns of the HEV genome. Taken together, synonymous codons with stable usage patterns in ORF1 might help to sustain the infection, while that with varied usage patterns in ORF3 may facilitate cross-species infection and expand the host range.
Abbreviations
-
- HEV
-
- hepatitis E virus
-
- ORF
-
- open reading frame
-
- RSCU
-
- relative synonymous codon usage
1 INTRODUCTION
Hepatitis E virus (HEV) is an important zoonotic pathogen. It has a single-stranded, positive RNA genome containing three open reading frames (ORFs). ORF1 produces a polyprotein consisting of nonstructural proteins, ORF2 expresses the capsid protein, and ORF3 encodes a small phosphoprotein with a multifunctional C-terminal region [1]. HEV has been classified into the Orthohepevirus genus of the Hepeviridae family [2]. According to taxonomy, currently available HEV strains are clustered into four species within the genus Orthohepevirus: Orthohepevirus A (strains derived from swine, wild boar, human, camel, rabbit, and ruminant), Orthohepevirus B (from avian), Orthohepevirus C (from rat and ferret), and Orthohepevirus D (from bat) [3]. Among the eight defined genotypes of HEV, genotypes 1 and 2 are generally limited to humans; whereas others. in particular, genotypes 3 and 4 infect various animal species [3]. HEV has an ever-expanding host range and these strains have high genetic diversity in nucleotide usages [4-6]. This genetic diversity is mainly attributed to the high mutation rate during viral replication catalyzed by RNA-dependent RNA polymerase [7] but also influenced by host factors.
Co-evolution between viruses and hosts is essential in shaping the expansion of the host range and cross-species infection of HEV [8-10]. However, the evolutionary dynamic interplay between HEV and different animal species in nature is difficult to recapitulate in laboratory-based studies that depend on a single viral clone infecting an isogenic host population. Therefore, the profile for evolutionary dynamics is critical for studying the spectrum of HEV infection influenced by the host factor. Among multiple evolutionary factors, the synonymous codon usage pattern is one of the bridges between viruses and hosts. A synonymous codon is redundant and most amino acids are encoded by more than one synonymous codon. The synonymous codon usage pattern is crucial in modulating the efficiency and accuracy of viral protein translation and maintaining the viral protein amino acid sequence [11-13]. Synonymous codon usages in the viral genome can reflect the active cross-talk between viral infection and host immune response [14-16]. Currently, there have been two major models to account for synonymous codon usage bias, namely the neutral (or mutational) model and the natural selective (or the translation-related selection) model. At the genomic or gene level, the mutational model has a strong correlation with genetic compositional constraints which can shape the probability of mutational fixation [17-20]. Most notably, it has been proved that elevated CpG/TpA dinucleotides content and enhanced host innate immune responses are also closely linked to synonymous codon usage [21-24]. In the natural selection model, the effects of synonymous codon usage patterns on translation accuracy/efficiency, messenger RNA half-life and secondary structure formation of protein have been intensively studied [25, 26]. It has been figured out that a positive correlation exists between transfer RNA (tRNA) abundance and synonymous codon usage patterns. As viruses do not have tRNAs, the translation of viral proteins would mainly depend on the pool of host tRNAs [27-29]. In this study, we have analyzed the dispersion of 59 synonymous codons in various HEV strains to understand the role of the host factor in shaping synonymous codon usages. Furthermore, we have investigated the evolutionary adaptation of HEV to its hosts, so as to identify the potential causes of the ongoing expansion of the host range and cross-species infection.
2 MATERIALS AND METHODS
2.1 HEV strains
The available complete genomes with ORF annotations of 297 HEV strains were downloaded from the National Center for Biotechnology. The demographics of the selected HEV strains were listed in Supporting Information: Table S1. Due to the limit of the amount of HEV strains isolated from wild deer, yak, goat, and cow, these strains were classified into one group as derived from the “ruminant” host.
2.2 Calculating the relative synonymous codon usage (RSCU) in HEV ORFs
The RSCU values for the three HEV ORFs were calculated without the negative influence of the amino acid compositions or the length of the coding sequence [30].
2.3 Analyzing the dispersion of synonymous codon usages in HEV ORFs derived from different hosts
Here, the synonymous codon pattern with Vs > 1.0 were considered as an obviously varied pattern of synonymous codon usage, while the synonymous codon usage with Vs < 1.0 were regarded as a stable pattern. Of note, when the synonymous codon usage with Vs < 0.1, the corresponding one was considered as a strong stable usage pattern.
2.4 Estimating the effect of the host on the overall codon usage patterns of HEV
3 RESULTS
3.1 Different synonymous codon usage patterns among the three HEV ORFs
The Vs values for 59 synonymous codons in the three HEV ORFs were calculated (Supporting Information: Tables S2–S4). Generally, the dispersion of 59 synonymous codons in HEV ORF1 displayed a relatively stable usage pattern, due to all synonymous codons with Vs < 1.0 (Supporting Information: Table S2). Interestingly, the synonymous codon usage pattern in HEV ORF1 derived from ruminants was significantly more stable (Vs < 0.1) than ones derived from humans, rabbits, and swine (Supporting Information: Table S2). Thus, the host factor, serving as one of the evolutionary dynamics, probably participates in synonymous codon usage variations in HEV ORF1.
The ruminant-derived HEV ORF2 had the largest number of synonymous codons with Vs < 0.1. However, these codons were rare in ORF2 from other hosts and even absent in ORF2 from avians (Supporting Information: Table S3). Of note, AGA coding for Arg never appeared in ORF2 derived from ruminant but was widely used in ORF2 originated from other hosts. The synonymous codon usage patterns in ORF3, compared to ORFs 1 and 2, were variable, but some specific synonymous codons were absent in ORF3 (Supporting Information: Table S4). Although ORF2 partially overlaps ORF3, many synonymous codons selected by ORF2 were not present in ORF3, further implying that evolutionary dynamics related to the host factor might partially shape genetic features of ORFs 2 and 3 derived from different hosts.
3.2 Species-specific usage pattern of synonymous codons in HEV ORFs
According to the Vs values of 59 synonymous codons in Supporting Information: Table S2–S4, the three ORFs of HEV derived from ruminants had the most synonymous codons with Vs < 0.1, while strains from humans had the fewest synonymous codons with Vs < 0.1 (Table 1). Generally, ORF1 contained more synonymous codons with Vs < 0.1 than ORFs 2 and 3. In other words, ORF1 derived from hosts excluding camel did not contain any synonymous codons with Vs > 1.0 and taking the synonymous codons (AGA or AGG) for Arg with Vs > 1.0 as an example, they were found in ORF2 from most animals (Table 2). Interestingly, ORF3 contained more synonymous codons with Vs > 1.0 than ORFs 1 and 2 (Figure 1). Overall, ORFs 1 and 2 shared a relatively stable usage pattern of synonymous codons (Vs < 1.0), while ORF3 had a variable one. Thus, synonymous codon usage patterns in ORFs indicate the contribution of the host factor to the evolutionary pathway of HEV.
| ORF1 | ORF2 | ORF3 | |
|---|---|---|---|
| Avian | GUG and GUU (Val), CAU and CAC (His), GCC and GCU (Ala), GAG (Glu), GGU (Gly), CGU (Arg), CCG and CCC (Pro), GAU and GAC (Asp), UUU (Phe) | ----- | CCG and CCC (Pro), GAU (Asp), CAG (Gln), GGC, GGG and GGA (Gly), UGC and UGU (Cys), AAC (Asn) |
| Camel | UUG (Leu), GUC (Val), CAG (Gln), AUU and AUC (Ile), CCU and CCA (Pro), CGG (Arg), AAU (Asn) | AUU (Ile), GGC and GGU (Gly) | AUA, AUU and AUC (Ile), GUU and GUA (Val), CCG (Pro), UCG (Ser) |
| Ferret | AUA (Ile), UAC and UAU (Tyr), CAG (Gln), CGG, CGU, CGC, CGA and AGG (Arg), GGG and GGC (Gly), AAU and AAC (Asn), CUC, CUU, CUG, UUG, UUA and CUA (Leu), GCC and GCG (Ala), ACU and ACA (Thr), GAG (Glu), UUU and UUC (Phe), GAU (Asp), UCU and UCC (Ser), UGU and UGC (Cys), CCU (Pro), AAG (Lys), GUG (Val) | UAC and UAU (Tyr), CAG and CAA (Gln), GGG (Gly), CGU, CGC and CGA (Arg), CUC and UUG (Leu), GCU and GCC (Ala), GAG (Glu), UCU, UCC and UCG (Ser), GAU and GAC (Asp), ACA (Thr), GUU (Val), AAG (Lys) | AGA, CGU and CGC (Arg), UCU and UCC (Ser), ACC (Thr), UGU and UGC (Cys), GUU and GUC (Val), AGC and UCA (Ser), CCG and CCC (Pro), AUC (Ile), CAC (His) |
| Human | GAG (Glu), CAG (Gln), GUU (Val), CCU (Pro), GCU (Ala) | GAG (Glu), CAG (Gln) | ----- |
| Rabbit | CAC and CAU (His), GAU and GAC (Asp), UCU (Ser) | GAU (Asp), CAG (Gln), CCC (Pro), GGG (Gly), AAU (Asn), GAA (Glu), GCU (Ala) | GGC (Gly), GAG (Glu) |
| Ruminant | CUU, CUC and CUG (Leu), AUU and AUC (Ile), GUU, GUC and GUG (Val), UCU, UCC, UCA, UCG and AGU (Ser), ACA and ACG (Thr), UGC (Cys), CGU, CGC, CGG and AGG (Arg), all synonymous codons for Phe, Pro, Ala, His, Gln, Asn, Lys, Asp, Glu, and Gly | UUG, CUU and CUA (Leu), AUU and AUC (Ile), GUU and GUC (Val), UCU, UCC, UCA, UCG and AGU (Ser), ACU, ACC and ACA (Thr), GCU and GCC (Ala), AAU (Asn), AAG (Lys), GAG (Glu), CGU and CGG (Arg), GGU and GGC (Gly), all synonymous codons for Phe, Pro, Tyr, His, Gln, and Asp | CAA (Gln), GCU and GCC (Ala), GUC (Val), UCG (Ser), CCU and CCG (Pro), ACC (Thr), GAU (Asp), GAG (Glu), CGU (Arg), GGC and GGG (Gly), all synonymous codons for Ile |
| Swine | GAG (Glu), CAG (Gln), AAG (Lys), GGG and GGC (Gly), GCU and GCC (Ala) | CAG (Gln) | GGG (Gly), AUU (Ile) |
| Wild boar | AAG (Lys), GAG (Glu), UUU (Phe), CAG (Gln), GGC (Gly), AUU (Ile), GCC (Ala), CGC (Arg) | GCU (Ala) | GGG (Gly) |
| Wild rat | GUG (Val), GAG (Glu), AAU and AAC (Asn), CAG (Gln), GGC and GGG (Gly), AAG (Lys), CCG (Pro), CUG (Leu), AGG (Arg), GCC (Ala), UAU (Tyr) | GAG (Glu), CAG (Gln) | ----- |
- Abbreviations: HEV, hepatitis E virus; ORF, open reading frames.
| ORF1 | ORF2 | ORF3 | |
|---|---|---|---|
| Avian | ----- | ----- | UUU (Phe), UAU (Tyr), CUU (Leu), CCU (Pro), AUU (Ile), AGG and CGA (Arg) |
| Camel | AGA (Arg) | AGA (Arg) | AAC (Asn), GAG (Glu), GCU (Ala), AGG and CGA (Arg), UAU (Tyr), UUU (Phe), UAC (Tyr), AGU (Ser), ACG (Thr), GGA (Gly) |
| Ferret | ----- | ----- | AUA (Ile), UAC (Tyr), ACU (Thr), UUU (Phe), CGA (Arg), GAC (Asp), GAA (Glu), AGU (Ser) |
| Human | ----- | AGA (Arg) | AAG (Lys), AGG and CGA (Arg), GGU (Gly), UAU and UAC (Tyr), ACU and ACG (Thr), GAA (Glu), UCA (Ser), GUA (Val) |
| Rabbit | ----- | AGA (Arg) | GAC (Asp), AGU and UCA (Ser), GUA (Val), AAU (Asn), GCU (Ala), UUA (Leu), GGA (Gly), CGA (Arg) |
| Ruminant | ----- | AGG (Arg) | AAC (Asn), UCA and AGU (Ser), AAG (Lys), GGA (Gly), GUA (Val) |
| Swine | ----- | AGA (Arg) | GUA (Val), CGA and AGG (Arg), ACG (Thr), UCA (Ser), GAA (Glu), UAU and UAC (Tyr), CAU (His), GGU (Gly) |
| Wild boar | ----- | AGA (Arg) | CAU (His), GGU (Gly), GUA (Val), GAA (Glu), UAC and UAU (Tyr), ACU and ACG (Thr), AGG and AGA (Arg) |
| Wild rat | ----- | AGA (Arg) | GGU (Gly), CAU (His), GUC and GUA (Val), GAC (Asp), UCC and UCA (Ser), CGA (Arg), ACG and ACU (Thr), AAA (Lys) |
- Abbreviations: HEV, hepatitis E virus; ORF, open reading frames.
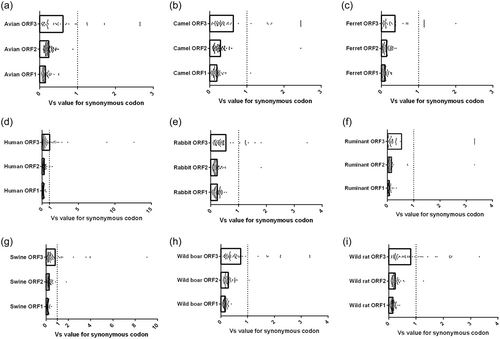
3.3 Variation of synonymous codon usage patterns in HEV ORFs
Information entropy was used to quantify the dispersions of overall codon usages of HEV derived from different hosts. As shown in Figure 2a, HEV derived from rabbits had the greatest dispersion of overall codon usages and HEV derived from ruminants had the smallest. HEV strains isolated from avian, camel, ferret, wild boar and wild rat displayed a relatively large dispersion of overall codon usages. The swine-derived HEV displayed a more stable usage pattern of synonymous codon than that isolated from wild boar. Interestingly, the human-derived HEV had a significantly different overall codon usage pattern compared with those from other hosts (Figure 2a).
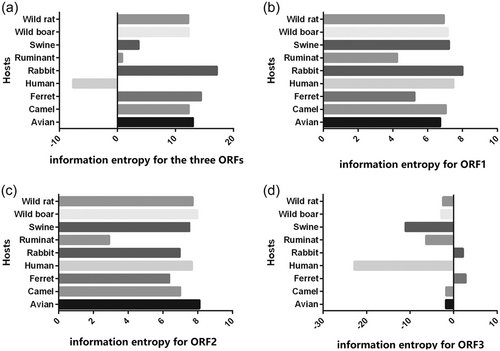
To better understand the contribution of different ORFs of HEV to the overall dispersion of synonymous codon usages, information entropy was used to analyze each ORF. As shown, the synonymous codon usage pattern of ORF3 was quite different from ORFs 1 and 2, and a similar relationship was observed between ORF1 and ORF2 (Figure 2b–d). Interestingly, the dispersions in HEV ORF3 derived from humans, swine, and ruminants were more varied than strains derived from other hosts (Figure 2d). These results reflect that the varied patterns of synonymous codon usage in ORF3 probably make more contribution to the overall dispersion for HEV derived from human, swine, and ruminants at synonymous codon usages.
4 DISCUSSION
In this study, we have analyzed the dispersion of synonymous codon usage patterns of HEV genomes derived from different hosts. Previous studies have pointed out the effects of a high mutation rate on synonymous codon usage bias of HEV [31, 32]. By using a different analysis workflow, we revealed that the host factor, serving as one of the evolutionary dynamics, plays an important role in the genetic divergence of HEV derived from different hosts at synonymous codon usages.
Synonymous codon usage bias, which can reflect the equilibrium between mutation pressure and natural selection, is a widespread genetic feature throughout the genomic evolution [33-35]. Although the HEV genome has a high mutation rate in its nucleotide sequence like other RNA viruses, the dispersion of the overall codon usage pattern of HEV displays the species-specific genetic feature. Based on the divergence of synonymous codon usage dispersions in HEV ORFs (Table 1), the evolutionary dynamic by host factor could participate in the overall codon usage pattern of HEV. Furthermore, considering those synonymous codons with Vs > 1.0 (Table 2), the synonymous codon usage variations with high frequencies are likely correlated with host range expansion of HEV infection. It has been suggested that the low synonymous codon usage bias of RNA viruses is an advantage for their efficient replication by reducing the competition between the virus and the host for the synthesis machinery. As RNA viruses can adopt various pathways to occupy the maximum utility from their informationally limited genome, RNA viruses co-evolved with their susceptible hosts that underwent a major evolutionary processing from synonymous codon usage variations [36-39]. ORF1 is the longest ORF in the HEV genome which is expressed directly from the viral genome to synthesize a polyprotein including methyltransferase, Y domain, papain-like cysteine protease, hypervariable region X domain, Hel and RNA-dependent RNA polymerase [40]. Compared with HEV ORFs 2 and 3, ORF1 has a more stable usage pattern of synonymous codon across various hosts (Figure 2). This suggests that the synonymous codon usage patterns of ORF1 are essential in the maintenance of the HEV life cycle in different hosts. Nevertheless, viral evolutional plasticity appears to limit the incorporation of new evolutional information with which to achieve more favorable host manipulation [36]. As ORF1 produces multiple viral nonstructural proteins which sustain viral replication and evade host immune defense [41], the evolution of a relatively stable synonymous codon usage pattern may benefit the infection and transmission. Interestingly, some previous studies pointed out that the processing of polyprotein translated by ORF1 exhibited different patterns [42-47], further implying that HEV ORF1 is likely processed in a fastidious manner and requires an optimum cellular condition that can sustain the relatively stable usage patterns of synonymous codons.
HEV ORF3 can frequently persist in immunocompromised hosts and facilitate viral egress in vitro [48, 49]. Although HEV ORF3 largely overlaps ORF2, the synonymous codon usage varies substantially in ORF3 (Figure 1). Co-evolution between a virus and its host is considered a positive selection between viral mutation and host immune defense [50]. Interaction between ORF3 and host proteins is essential for the release of HEV particles from the infected cells [51, 52]. These diverse biological functions of ORF3 may be reflected by its genetic divergence of synonymous codon usage patterns. Importantly, the synonymous codon usage dispersion of ORF3 substantially contributes to the overall synonymous codon usage patterns in HEV genomes originating from different hosts (Figure 2). The expression level of ORF3 is related to the severity and prognosis of HEV infection and ORF3 protein is considered a sensitive and specific diagnostic index [53]. The synonymous codon usage patterns of ORF3 derived from different hosts may thus regulate the expression level in target cells [16, 54, 55]. The synonymous codon usages of different ORFs display divergence according to the origin of the host species. These genetic features suggest that high mutation rates in the HEV genome might occur in a limited rather than permissive manner. The consequent selection of mutations in the RNA virus genome likely favors viral adaptation to its susceptible hosts [56]. Synonymous codon usage variations of the HEV genome may be a genetic result that enables the virus to survive in a specific host and to achieve cross-species infection. Generally, HEV isolated from humans has a divergent pattern of the overall codon usage in comparison with the ones from other hosts. The codon usage biases of ORFs between genotype 1 and genotypes 3 and 4 might reflect the role of ORF1 and ORF2 in shaping human-restricted HEV and zoonotic HEV. However, the divergence of codon usage bias in ORF2 between genotype 1 and other genotypes is weaker than that in ORF1 between genotype 1 and other genotypes. The previous studies pointed out ORF2 gene is not involved in HEV cross-species infection, by means of inserting ORF2 of the zoonotic genotypes 3 and 4 HEV into the genomic backbone of a genotype 1 human HEV failed to infect pigs [57, 58]. However, when the ORF1 of genotype 4 HEV was inserted into the backbone of a genotype 1 HEV, this intergenotypic chimeric virus altered its host cell tropism and infects pig kidney cell line, further suggesting a potential role of ORF1 in HEV cross-species infection [59]. Based on these previous studies, natural selection derived from humans plays an important role in shaping codon usage of human-derived HEV. Moreover, one previous study, which investigated the genetic feature of synonymous codon usages between human-derived HEV and swine-derived HEV, suggested that high synonymous codon variations are likely correlated with host range expansion of HEV infection [60]. Taken together, natural selection derived from the specific host probably participates in shaping synonymous codon usages of HEV to adapt to the corresponding host.
We found that the host factor strongly contributes to the genetic divergence of HEV derived from diverse hosts. The huge dispersion of synonymous codon usage in ORF3 may partially mediate the expansion of host range and cross-species infection of HEV. However, it remains a challenge to fully understand the ever-expanding host range and cross-species infection of HEV, due to the lack of sufficient knowledge of evolutionary dynamics related host factors. Future studies exploiting genetic divergence between closely related HEV strains will help to reveal the fundamental rules that govern co-evolution between hosts and HEV populations.
ACKNOWLEDGMENTS
This study was supported by the Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital (CY2019-MS07 and CY2021-BJ-A18), Innovation Foundation for Higher Education Institution of Gansu Province (2020B-022), and Gansu Province Science and Technology Fund (20JR10RA737).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




