Neutralization of SARS-CoV-2 Omicron XBB.1.5 and JN.1 variants after COVID-19 booster-vaccination and infection
Karin Stiasny and Judith H. Aberle contributed equally
Abstract
SARS-CoV-2 Omicron lineages continue to emerge and evolve into new sublineages, causing infection waves throughout 2022 and 2023, which has been attributed to immune escape. We examined neutralizing antibody responses to the recently emerged SARS-CoV-2 JN.1 variant in comparison to ancestral D614G and Omicron BA.1, BA.2, BA.5, and XBB.1.5 variants. We tested 79 human sera from cohorts with different combinations of vaccinations and infections, including 23 individuals who had been repeatedly exposed to Omicron. Individuals with a monovalent XBB.1.5 vaccine booster or XBB.1.5 breakthrough infection had robust antibody levels against all variants tested; however, JN.1 evaded antibodies in individuals after single Omicron BA.1, BA.2 or BA.5 breakthrough infections. Moreover, in the non-vaccinated cohort, serum antibodies demonstrated almost no cross-neutralization activities against D614G, XBB.1.5 and JN.1. after infections with earlier Omicron variants. These findings show that SARS-CoV-2-immunity is heterogeneous, depending on different combinations of vaccinations and infections, and emphasize the importance of considering different immune-backgrounds when evaluating novel variants.
1 INTRODUCTION
SARS-CoV-2 Omicron variants continue to evolve into multiple sublineages that can evade immunity due to mutations in its spike protein.1, 2 This has caused global waves of infections with novel Omicron lineages that escape immunity from infection or vaccination, requiring vaccine adaptations. Bivalent mRNA vaccines that target both ancestral (D614G) and Omicron BA.1 or BA.5 spike proteins were used as booster vaccines, starting in September 2022. In September 2023, a monovalent Omicron XBB.1.5 vaccine was introduced to elicit neutralizing antibodies against current Omicron variants. The ongoing encounters with various SARS-CoV-2 variants and combinations of different vaccine types and infections led to multiple diverse immune backgrounds in the population and present challenges for vaccination strategies and risk assessment concerning the behavior of novel variants.
New Omicron sublineages, including those that have descended from BA.2 or BA.5 variants have emerged, including BQ.1, XBB.1.5, EG.1, as well as BA.2.86 and its descendent JN.1, with the latter two containing more than 30 spike mutations relative to its parental BA.2 variant.3 Recent studies indicate that infection with BA.2.86 or JN.1 results in higher rates of viral shedding compared to XBB.1.5, including in presymptomatic and vaccinated individuals, but there is no evidence for different symptom profiles or an increased disease severity.4, 5 In comparison with BA.2.86, JN.1 accumulated an additional mutation (L455S) in the receptor-binding domain of the spike protein, suggesting enhanced immune escape properties. Although early reports have shown that neutralizing antibody responses to BA.2.86 and JN.1 were comparable or slightly lower than to other recently circulating variants,6-10 the JN.1 variant, in contrast to BA.2.86, increased rapidly in frequency according to GISAID sequences in late 2023.3 It quickly became dominant worldwide, replacing earlier variants, and was the main lineage causing the epidemic wave in December 2023–January 2024, leading to its classification as a separate variant of interest by the World Health Organization. In this study, we evaluated serum neutralizing antibodies with live-virus neutralization assays with the SARS-CoV-2 ancestral (D614G) strain as well as the Omicron BA.1, BA.2, BA.5, XBB.15 and JN.1 variants. To assess potential immune escape by JN.1, we analyzed neutralization titers of cohorts with diverse vaccination and Omicron infection histories.
2 METHODS
2.1 Omicron variant identification
Nasopharyngeal swabs from routine diagnostics were PCR-tested for SARS-CoV-2. Variants were identified using Vir-SNiP SARS-CoV-2 assays (TIB MOLBIOL) to distinguish between BA.1, BA.2, BA.5 and XBB.1.5. In some cases, variants were additionally confirmed by next-generation sequencing using amplification with ARTIC network tiled amplicon primers followed by Nextera XT library preparation and sequencing on an Illumina MiSeq device.
2.2 Live-virus SARS-CoV-2 neutralization test (NT)
SARS-CoV-2 strains were amplified on VeroE6 (ECACC #85020206) or VeroE6-TMPRSS2 cells (courtesy of Anna Ohradanova-Repic), as described previously.11-14 Sequences determined by next-generation sequencing were uploaded to the GISAID database (Supporting Information S1: Table S1).13
Live-virus NTs were conducted as described earlier.11-14 In brief, 50-100 TCID50 SARS-CoV-2 were incubated with serial two-fold dilutions from 1:10 up to 1:20,480 of heat-inactivated serum (in duplicates) for 1 h at 37°C. The sample-virus mixture was added to VeroE6 cells and incubated for 3–5 days at 37°C. Cytopathic effects (CPE) was microscopically assessed. Neutralizing titers were determined as the last reciprocal dilution that prevented CPE.
2.3 Serum samples
The study was conducted in accordance with the Declaration of Helsinki. We used anonymized leftover samples from diagnostic antibody testing. A total of 79 serum samples was included. Of these, 59 were obtained from vaccinated individuals who had subsequently experienced an Omicron (BA.1, BA.2 or BA.5) infection, and 20 samples were obtained from individuals who had one or two Omicron infections without prior vaccination. Leftover samples were stored in the biobank of the Center for Virology, following established protocols (EK1513/2016) in accordance with national legislation, that required no additional written consent. The ethics committee of the Medical University of Vienna, Austria approved the study protocol (EK 1291/2021).
2.4 Statistical analyses
Statistical analysis and data visualization were performed with R version 4.2.0 and Adobe Illustrator.
3 RESULTS
We analyzed serum samples from cohorts who had previously received three to four doses of an ancestral vaccine, and had at least one infection with Omicron variants, as detailed in the Supplementary Table 2. Four different constellations of SARS-CoV-2 immunity were defined: boosted with the monovalent XBB.1.5 vaccine (n = 8); infected with Omicron XBB.1.5 (n = 10); infected with Omicron BA.5 (n = 4); or boosted with the bivalent BA.5 vaccine (n = 9). As reference cohorts, we included samples from individuals who had received three to four vaccine doses, followed by infection with Omicron variants BA.1 (n = 11), BA.2 (n = 7) or BA.5 (n = 10). Additionally, we included a group of unvaccinated individuals with Omicron BA.5 infection with or without a previous Omicron BA.1 or BA.2 infection (n = 20). Details regarding demographic and clinical characteristics of the study population are provided in Supporting Information S1: Tables 2–4. Serum neutralizing antibody titers were determined against six SARS-CoV-2 variants (D614G, Omicron BA.1, BA.2, BA.5, XBB.1.5, and JN.1) using a live-virus neutralization assay, as previously described.11-14
We found that serum samples 1-2 months after XBB.1.5 vaccination (Omicron inf + XBB.1.5 Vac) or XBB.1.5 breakthrough infection (Omicron inf + XBB.1.5 inf) displayed high neutralizing capacity against all variants tested (Figure 1A,B). In both cohorts, the serum titers against XBB.1.5 and JN.1 differed by a factor of 2.2–2.4, and were approximately 2 to 7-fold lower than those against D614G. Reduced neutralization of JN.1 was also observed 1–2 months after an Omicron BA.5 breakthrough infection in individuals with a previous Omicron BA.1 or BA.2 infection (Omicron inf + BA.5 inf) (Figure 1C). Neutralizing antibody titers against JN.1 were three-fold lower than those against XBB.1.5, and 14.5-fold lower than those against D614G (Figure 1C). A considerably stronger immune escape by JN.1 was detected in individuals after bivalent BA.5 vaccination (Omicron inf + Biv BA.5 Vac) (Figure 1D). Notably, neutralization titers against JN.1 were 13.5-fold lower than those against XBB.1.5 and 29.8-fold lower than those to D614G (Figure 1D).
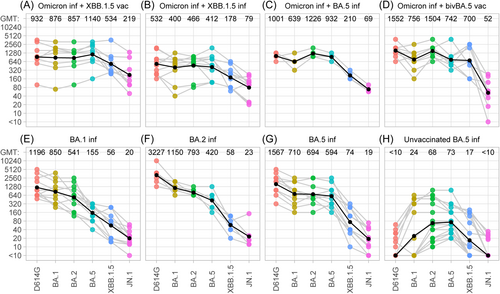
We also analyzed serum titers of previously reported cohorts who had a single Omicron breakthrough infection without an updated vaccine booster. Antibody titers in these samples were determined against JN.1, and neutralization data for D614G, BA.1, BA.2, BA.5 and XBB.1.5 were extracted from our previous report.13 The three Omicron breakthrough cohorts exhibited a strong immune escape by both, JN.1 and XBB.1.5, with serum titers that were 21- to 140-fold lower than those against D614G (Figure 1E–G). Finally, we measured neutralization after one or two Omicron infections of non-vaccinated individuals. These sera neutralized Omicron BA.2 and BA.5, whereas cross-neutralization against D614G, BA.1, XBB.1.5 was detected in 35%, 40% and 60%, respectively, and in only 25% against JN.1 (Figure 1H).
4 DISCUSSION
Our findings demonstrate that the neutralization capacity against newly emerging Omicron variants is very heterogeneous and strongly depends on the infection and vaccination history of an individual. In the case of the currently prevalent JN.1 variant, potent neutralization was observed for vaccinated persons with sequential Omicron exposures, while those after single Omicron breakthrough infections without an updated vaccine boost, or repeated Omicron infections in the absence of vaccination demonstrated little or no cross-neutralizing activities against JN.1. In particular, we showed that sera after XBB.1.5 booster vaccination or breakthrough infection showed robust neutralization against all variants tested, although titers were approximately two-fold lower for XBB.1.5 than those for D614G and further reduced for JN.1 by 2.2- to 2.4-fold. These findings are in agreement with recent experimental studies, which observed 2.9- to 4.3-fold decreased neutralization titers against JN.1 relative to XBB.1.5 after an XBB.1.5 monovalent mRNA vaccine booster.7 Moreover, epidemiological data reported a lower level of protection against BA.2.86 and JN.1 infections by XBB.1.5-vaccine-induced antibodies, although the differences were not significant.15, 16
Here, we extended our study to cohorts with different infection and vaccination histories, including individuals with a bivalent BA.5 vaccine booster or BA.5 breakthrough infections after a prior Omicron BA.1 or BA.2 infection. Notably, we observed an enhanced immune escape by JN.1 for bivalent vaccine-boosted individuals who showed 30-fold lower titers than those against D614G. A considerable reduction in JN.1 titers was also observed in individuals who had a BA.5 breakthrough infection following a previous Omicron BA.1 or BA.2 infection, however, the reduction in titers against JN.1 over D614G was not as large (14.5-fold) as observed for the bivalent BA.5 vaccine. These findings are in line with previous studies, demonstrating a strong back-boosting to D614G by the ancestral spike included in bivalent vaccines,17 resulting in increased titers against D614G. Importantly, despite the visible fold-reduction in titers relative to D614G, neutralization against XBB.1.5 and JN.1 was observed in almost all individuals of the bivalent BA.5 vaccine-boosted cohort and vaccinated individuals with sequential Omicron infections. In contrast, low or undetectable neutralization against JN.1 was observed in vaccinated individuals who had a single Omicron breakthrough infection, as well as in individuals who were not vaccinated and experienced one or two Omicron infections. Moreover, in the non-vaccinated cohort, serum antibodies demonstrated almost no cross-neutralization activities against D614G, XBB.1.5 and JN.1. These findings are in agreement with studies using Omicron BA.1 and BA.2 postinfection samples from non-vaccinated persons,9, 12 as well as neutralization data from mice primed with Omicron BA.5 18 and emphasize the importance of booster vaccination with updated vaccines even in populations with previous Omicron infections. The study limitations included the small sample numbers, and that we evaluated antibody responses only at approximately 1 month after vaccination or breakthrough infection and did not address how such responses evolve over time.
In summary, our results showed that heterogeneous humoral immunity against newly emerging SARS-CoV-2 lineages, such as Omicron JN.1, leads to diverse immune evasion properties, dependent on different combinations of vaccinations and infections.
ETHICS APPROVAL
The study was conducted in accordance with the Declaration of Helsinki. We used anonymized leftover samples from diagnostic antibody testing, stored in the biobank of the Center for Virology, following established protocols (EK1513/2016) in accordance with national legislation, that required no additional written consent. The ethics committee of the Medical University of Vienna, Austria approved the study protocol (EK 1291/2021). The study was performed at the Center for Virology of the Medical University of Vienna.
AUTHOR CONTRIBUTIONS
David N. Springer: Conceptualization; writing of the manuscript; methodology and experimental work; data analysis. Jeremy V. Camp: Methodology and experimental work. Stephan W. Aberle: Methodology and experimental work. Lukas Weseslindtner: Methodology and experimental work. Karin Stiasny: Writing of the manuscript; methodology and experimental work; resources; data analysis; supervision. Judith H. Aberle: Conceptualization; writing of the manuscript; resources; data analysis; funding acquisition; supervision. All authors contributed to the manuscript and approved the submitted version.
ACKNOWLEDGEMENTS
We thank Jutta Hutecek, Sylvia Malik, Barbara Dalmatiner for their excellent technical assistance. This work was supported by the Medical-Scientific fund of the Mayor of the federal capital Vienna (grant 23096).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The datasets generated and/or analyzed during the current study are available from the corresponding authors on reasonable request.




