Investigating the role of killer cell immunoglobulin-like receptors and human leukocyte antigen genetic variants in hepatitis C virus infection
Yuwen Li, Tian Zeng, and Peng Huang contributed equally to this study.
Abstract
The genetic diversity of killer cell immunoglobulin-like receptors (KIRs) and human leukocyte antigen (HLA) genes influences the host's immune response to viral pathogens. This study aims to explore the impact of five single nucleotide polymorphisms (SNPs) in KIR3DL2 and HLA-A genes on hepatitis C virus (HCV) infection. A total of 2251 individuals were included in the case-control study. SNPs including KIR3DL2 rs11672983, rs3745902, rs1654644, and HLA-A rs3869062, rs12202296 were genotyped. By controlling various confounding factors using a modified logistic regression model, as well as incorporating stratified analysis, joint effects analysis, and multidimensional bioinformatics analysis, we analyzed the relationship between SNPs and HCV infection. The logistic regression analysis showed a correlation between KIR3DL2 rs11672983 AA, KIR3DL2 rs3745902 TT, and increased HCV susceptibility (p < 0.01). Stratified analysis indicated that KIR3DL2 rs1654644 and HLA-A rs3869062 also heightened HCV susceptibility in certain subgroups. A linear trend of rising HCV infection rates was observed when combining KIR3DL2 rs11672983 AA and KIR3DL2 rs3745902 TT (ptrend = 0.007). Bioinformatics analysis suggested these SNPs' regulatory potential and their role in altering messenger RNA secondary structure, implying their functional relevance in HCV susceptibility. Our findings indicate that KIR3DL2 rs11672983 AA and KIR3DL2 rs3745902 TT are significantly associated with increased susceptibility to HCV infection.
1 INTRODUCTION
Globally, an estimated 70 million individuals grapple with the enduring affliction of hepatitis C virus (HCV) infection.1, 2 Direct-acting antiviral drugs (DAAs) have demonstrated remarkable efficacy in the treatment of Hepatitis C, not just in the general populace, but also in high-risk groups where they exhibit high rates of cure.3-5 However, the lack of a durable immunological defense post-treatment leaves individuals vulnerable to reinfection.6 The determinants of viral clearance and sustained protective immunity remain unclear.7 In addition to the lack of an effective vaccine, achieving the World Health Organization's goal of eliminating hepatitis C by 2030 remains a significant global challenge.8 In this context, it is necessary to conduct in-depth research on the genetic factors associated with hepatitis C infection through large-scale population studies.
It is well known that the interaction between the host's immune responses and the virus plays a pivotal role in shaping the course and outcome of infection.9 Natural killer (NK) cells are a crucial component of the innate immune system, playing a key role in preventing and controlling infections, as well as recognizing and eliminating abnormal cells.10 They serve as frontline defense in the immune system. In HCV infection, NK cells not only have the ability to directly kill HCV-infected liver cells but also play a role in regulating adaptive immune responses, inhibiting virus replication, and activating other immune cells to participate in antiviral defense.11 The normal execution of these biological functions of NK cells relies on intricate receptor-ligand interactions within the immune system. Especially noteworthy is the collaborative interaction between killer cell immunoglobulin-like receptors (KIR) on NK cells and their ligands, the human leukocyte antigen (HLA) class I molecules, serving as the cornerstone in modulating NK cell activity.12 KIRs fine-tune the reactivity of NK cells by engaging with HLA class I molecules on target cells, oscillating between inhibition and activation.13, 14 Under pathological conditions, any aberrant factors influencing the expression of KIRs and their corresponding ligands, HLA class I, or their abnormal binding, may potentially lead to the dysregulation of NK cell functions.15 It is evident that the proteins encoded by KIR/HLA genes, along with the variations among these genes, play a crucial regulatory role in the immune system. The host genetic heterogeneity within KIR/HLA class I genes plays a crucial role in shaping the immune response to a range of viral pathogens.
KIR3DL2 is one of the “framework genes” in the KIR gene family, universally present in diverse human populations, with HLA-A being one of its ligands.16 KIR3DL2 is composed of three immunoglobulin-like domains—D0, D1, and D2. The D0 domain binds to class I HLA molecules, enhancing receptor-ligand interaction and fine-tuning the regulatory mechanism of NK cell signaling.17 HLA-A is widely expressed on the surface of eukaryotic cells, consisting of a heavy α chain and a β 2-microglobulin non-covalently bound together. Its function is to present endogenous epitopes derived from viral invasion or tumor cell aberrations to CD8+ T cells, triggering T cell-mediated immune responses against damaged cells.18 The investigative efforts led by Morgane et al. demonstrates that the expression of KIR3DL2 is associated with acute-type Adult T-cell leukemia, and can be triggered by infection with human T-cell leukemia virus type 1.19 The research conducted by Wu et al. has delineated a correlation between variations in KIR3DL2 and the emergence of certain cancers.20 Lunemann et al. discerned that KIR3DS1 and its cognate ligand HLA-F undergo upregulation within HCV-infected cellular environments, facilitating NK cell-mediated containment of HCV.21 The study by Sakhaee et al. found that HLA rs4273729 can serve as a potent predictor for HCV rapid virological response and Sustained Virological Response (SVR).22 In our previous studies, we found that the single nucleotide polymorphisms (SNPs) of HLA-DP and HLA-DP, KIR2DS4 rs35440472 and HLA-C rs1130838, as well as KIR2DL4 rs660773 and HLA-G rs9380142, are also associated with increased susceptibility to HCV infection.23-27 Therefore, based on the above findings and areas that remain unclear, this study aims to explore the genetic associations of five potential functional SNPs within the KIR3DL2/HLA-A and HCV infection. The candidate SNPs in our study including KIR3DL2 rs11672983, KIR3DL2 rs3745902, KIR3DL2 rs1654644, HLA-A rs3869062, and HLA-A rs12202296.
To avoid potential confounding effects from known genetic backgrounds, we included interferon lambda 4 (IFNL4) in our considerations, which has been confirmed to be closely associated with HCV infection and the response to treatment.28 Specifically, IFNL4 rs12979860 and IFNL4 rs8099917 are significantly associated with spontaneous virus clearance and response to peg-IFN-a/RBV treatment.29 Therefore, these two SNPs were also included in the study, and their genetic typing results were incorporated as confounding factors in the final analysis to avoid the possible effect of the proven IFNL4 genotype on the HCV infection outcome. We expect that this study will provide new insights into the host factors influencing the immune response to HCV infection.
2 MATERIALS AND METHODS
2.1 Subjects
A total of 2251 individuals at high risk of HCV infection were recruited from Jiangsu Province, China. The participants included 1682 paid blood donors from 20 villages and 569 dialysis patients (DP) from 9 hospitals. Data collection spanned the years 2011 to 2018. The inclusion criteria for the study are as follows: have not received DAAs or interferon drug treatment before enrollment and blood sample collection; follow-up for 6 months or more; signed informed consent form and voluntarily enrolled. Participants in the case group must test positive for HCV antibodies. The exclusion criteria for the study subjects are as follows: co-infection with human immunodeficiency virus or hepatitis B virus; ages below 18 or above 80 years; a prior history of anti-HCV treatment (using interferon or DAAs); and the presence of autoimmune, alcohol-induced, or metabolic liver disorders. The study subjects were divided into three groups based on the following criteria: Group A consisted of uninfected controls who tested negative for both serum anti-HCV antibodies and HCV RNA; Group B comprised individuals with a persistent infection, confirmed by positive results for both serum anti-HCV antibodies and HCV RNA; and Group C included those who achieved spontaneous viral clearance, as indicated by a positive anti-HCV antibody test but a negative HCV RNA result. Subjects from Groups B and C are collectively referred to as the “HCV infection group.” To ensure the accuracy of the serological results, each test was confirmed through at least three distinct serological assays over a 6-month observation period. Only specimens with consistent test results were included in the study. Comprehensive data for each subject, including demographic details, exposure to high-risk behaviors, laboratory findings, and a clinical profile of HCV infection, were collected using a meticulously designed questionnaire administered by experts in the field.
2.2 Serological testing
The study subjects' fasting venous blood (5−10 mL) was collected in EDTA anticoagulant tubes, followed by centrifugation to separate plasma and blood cells. The samples were then stored at −80°C for future use. HCV-specific antibodies were determined through quantitative enzyme-linked immunosorbent assay (Diagnostic Kit for Antibody to HCV 3.0 ELISA; Intec Products Inc.). HCV RNA levels were quantified using real-time fluorescence quantitative PCR, with a detection limit of 20 IU/mL (HCV Nucleic Acid Quantification Test Kit; Daan Gene Co., Ltd.). HCV genotypes were detected using the Murex HCV Serotyping Assay ELISA Kit (Abbott).30
2.3 Candidate SNP selection and genotyping
Candidate KIR3DL2/HLA-A tagSNP were retrieved from the 1000 Genomes Project SNP data set (37.0 version) and selected using the Haploview software (version 4.2). The selection criteria of tagSNP included: a minor allele frequency ≥ 0.05 and a pairwise linkage disequilibrium (LD) r^2 v ≥ 0.8 within the Chinese Han population. Besides this, screening the candidate SNP should be combined with previous literature reports. Previous studies have identified correlations between specific KIR3DL2/HLA-A SNPs and diseases: KIR3DL2 rs11672983 is linked to a gradual increase in the risk of preterm birth31; KIR3DL2 rs3745902 and KIR3DL2 rs1654644 are associated with pemphigus.32 HLA-A rs3869062 is connected to nasopharyngeal carcinoma,33 and HLA-A rs12202296 is associated with Stevens-Johnson syndrome and toxic epidermal necrolysis, both of which are potentially fatal drug eruptions related to T-cell-mediated immune dysregulation.34 Considering the factors mentioned above, the following five SNPs were finally chosen as candidate SNPs for the further study: KIR3DL2 rs11672983, KIR3DL2 rs3745902, KIR3DL2 rs1654644, HLA-A rs3869062, and HLA-A rs12202296. The basic information of candidate SNPs can be found in Supporting Information S1: Table 1 (Supporting Information S1: Table 1).
The TaqMan allele discrimination assay was executed on the Roche LightCycler® 480 II Real-Time PCR System.35 The primer and probe sequences for SNPs are detailed in the supplementary table 2 (Supporting Information S1: Table 2). The SNPs were genotyped in a blinded manner, with a success rate of over 90%. In addition, 10% of random samples were re-genotyped, yielding a 100% consistency rate. All tests were carried out in accordance with the manufacturer's instructions. The genotyping experiments of all samples used identical procedures and instruments. Genotyping data were interpreted using the LightCycler® 480 software (version 1.5.1).
2.4 In silico analysis
The functional implications of the SNPs were assessed with the aid of the SNP Function Prediction (FuncPred) tool, accessible at https://snpinfo.niehs.nih.gov/, and the HaploReg v4.2 database, found at https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php. The potential regulatory functions of these SNPs were also evaluated according to their RegulomeDB scores, accessible via the RegulomeDB online resource (http://www.regulomedb.org/). We utilized the Vienna RNA Web Services' RNAfold server (http://rna.tbi.univie.ac.at/) to forecast alterations in messenger RNA (mRNA) secondary structures attributable to SNP mutations. The University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/cgibin/hgGateway) served as a resource for investigating the potential biological functions of these SNPs. In addition, we utilized data from the Encyclopedia of DNA Elements (ENCODE) project to examine the expression levels of histone markers—histone H3 lysine 4 monomethylation (H3K4Me1) and lysine 27 acetylation (H3K27Ac)—across seven distinct cell lines (H1-hESC, HSMM, HUVEC, K562, NHEK, GM12878, and NHLF) to elucidate the role of SNPs in gene regulation and cell identity maintenance, as well as their functional differences across different cellular and biological contexts, to inform our analysis.
2.5 Statistical analysis
Conformity with the Hardy−Weinberg equilibrium (HWE) within the control cohort was verified using the chi-square (χ2) test. Variations in demographic, clinical, and virological data were compared using the χ2 test, one-way analysis of variance, or the Kruskal−Wallis test, as appropriate. Logistic regression adjusted for gender, age, route of infection, levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST), as well as genotypes of IFNL4 rs12979860 and IFNL4 rs8099917, was performed to analyze the association between candidate SNPs and HCV infection. by calculating odds ratio (OR) and 95% confidence interval (CI). Four different genetic models, including codominant, dominant, additive, and recessive frameworks, were employed to investigate the relationship between each SNP and the outcomes of HCV infection. Bonferroni correction was used to correct for multiple comparisons, and the p Value was adjusted to 0.01 (0.05/5). Statistical analyses were conducted using the SPSS software (version 26.0).
3 RESULTS
3.1 Demographic and clinical characteristics
The demographic and clinical characteristics of the participants are presented in Table 1. This study included a total of 2251 individuals, consisting of 1352 in Group A, 576 in Group B, and 323 in Group C. Significant differences were found in the distribution of gender, age, ALT, AST, infection route, and HCV genotype among the three groups (all p < 0.001). In the three cohorts, the gene phenotype distribution of IFNL4 rs12979860 and IFNL4 rs8099917 did not exhibit significant differences (all p > 0.05). The allele frequencies of the five candidate SNPs within control group conformed to the HWE (all p > 0.05).
| Variables | Group A n (%) n = 1352 |
Group B n (%) n = 576 |
Group C n (%) n = 323 |
p |
|---|---|---|---|---|
| Age (mean ± SD) | 58.72 ± 12.33 | 55.71 ± 9.19 | 57.95 ± 8.61 | |
| <50 | 316 (23.37) | 159 (27.6) | 48 (14.86) | <0.001a |
| ≥50 | 1036 (76.63) | 417 (72.4) | 275 (85.14) | |
| Gender | ||||
| Male | 513 (37.94) | 159 (27.6) | 99 (30.65) | <0.001a |
| Female | 839 (62.06) | 417 (72.4) | 224 (69.35) | |
| ALT (U/L) | ||||
| <50 | 1314 (97.91) | 392 (68.29) | 272 (84.47) | <0.001a |
| ≥50 | 28 (2.09) | 182 (31.71) | 50 (15.53) | |
| AST (U/L) | ||||
| <40 | 1283 (95.6) | 341 (59.41) | 253 (78.57) | <0.001a |
| ≥40 | 59 (4.4) | 233 (40.59) | 69 (21.43) | |
| Routes of infection | ||||
| DP | 416 (30.77) | 65 (11.28) | 88 (27.24) | <0.001a |
| PBD | 936 (69.23) | 511 (88.72) | 235 (72.76) | |
| HCV genotype | ||||
| 1 | - | 135 (38.35) | 42 (47.19) | <0.001b |
| Non-1 | - | 18 (5.11) | 28 (31.46) | |
| Mixed | - | 199 (56.53) | 19 (21.35) | |
| IFNL4-rs12979860 | ||||
| CC | 1142 (90.49) | 506 (89.24) | 282 (90.68) | 0.674a |
| CT/TT | 120 (9.51) | 61 (10.76) | 29 (9.32) | |
| IFNL4-rs8099917 | ||||
| TT | 1103 (87.96) | 502 (89.17) | 279 (89.42) | 0.648a |
| TG/GG | 151 (12.04) | 61 (10.83) | 33 (10.58) |
- Note: Non-1: HCV genotypes other than genotype 1, such as genotype 2, genotype 3, genotype 4, genotype 5, and genotype 6. Mixed: Coexistence of two or more HCV genotypes at the same time. Bold type indicates statistically significant results.
- Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; DP, dialysis population; Group A, uninfected control group; Group B, persistent infection group; Group C, spontaneous clearance group; HCV, hepatitis C virus; IFNL4, interferon lambda 4, also know as IL28B; PBD, paid blood donors.
- a p Value of χ2-test among three groups;
- b p Value of χ2-test among two groups.
3.2 Association of candidate SNPs with the susceptibility to HCV infection
Individuals from Groups B and C were combined into a unified HCV infection cohort for comparison with the uninfected control Group A to determine whether candidate KIR3DL2/HLA-A SNP were related to HCV susceptibility. The genotypic distribution of five SNPs is detailed in Table 2. After adjusting for confounding variables including age, gender, ALT, AST, IFNL4 rs12979860, and IFNL4 rs8099917, routes of infection, the results of logistic regression analysis revealed that individuals carrying the KIR3DL2 rs11672983 AA genotype and KIR3DL2 rs3745902 TT genotype exhibit an increased susceptibility to HCV infectiong compared to those with the rs11672983 wild-type GG genotype or rs3745902 wild-type CC genotype, respectively (KIR3DL2 rs11672983: adjusted OR = 1.630, 95% CI = 1.134−2.342, p = 0.008; KIR3DL2 rs3745902: adjusted OR = 1.692, 95% CI = 1.175−2.437, p = 0.005). Moreover, the analysis of the regression genetic model also revealed similar results (KIR3DL2 rs11672983: adjusted OR = 1.623, 95% CI = 1.142−2.308, p = 0.007; KIR3DL2 rs3745902: adjusted OR = 1.723, 95% CI = 1.210−2.453, p = 0.003). It is noteworthy that they remained signifcant after the multiple comparisons using Bonferroni correction.
| Gene | SNPs (genotype) | Group A n (%) n = 1352 |
Group B n (%) n = 576 |
Group C n (%) n = 323 |
Group (B + C) n (%) n = 899 |
OR (95% CI)a | pa | OR (95% CI)b | pb |
|---|---|---|---|---|---|---|---|---|---|
| KIR3DL2 | rs11672983 | ||||||||
| GG | 722 (54) | 299 (52.27) | 168 (52.01) | 467 (52.18) | 1 | 1 | |||
| GA | 529 (39.57) | 218 (38.11) | 126 (39.01) | 344 (38.44) | 1.009 (0.821-1.241) | 0.931 | 0.997 (0.729-1.363) | 0.983 | |
| AA | 86 (6.43) | 55 (9.62) | 29 (8.98) | 84 (9.39) | 1.630 (1.134-2.342) | 0.008 | 0.893 (0.530-1.504) | 0.67 | |
| Dominant model | 1.097 (0.902-1.334) | 0.355 | 0.975 (0.726-1.308) | 0.865 | |||||
| Recessive mode | 1.623 (1.142-2.308) | 0.007 | 0.894 (0.540-1.481) | 0.664 | |||||
| Additive model | 0.945 (0.774-1.155) | 0.583 | 1.014 (0.749-1.372) | 0.929 | |||||
| KIR3DL2 | rs3745902 | ||||||||
| CC | 706 (52.45) | 294 (51.31) | 165 (51.24) | 459 (51.28) | 1 | 1 | |||
| CT | 556 (41.31) | 222 (38.74) | 129 (40.06) | 351 (39.22) | 0.960 (0.781-1.180) | 0.697 | 1.008 (0.738-1.378) | 0.96 | |
| TT | 84 (6.24) | 57 (9.95) | 28 (8.7) | 85 (9.5) | 1.692 (1.175-2.437) | 0.005 | 0.834 (0.494-1.408) | 0.497 | |
| Dominant model | 1.056 (0.868-1.284) | 0.585 | 0.971 (0.723-1.304) | 0.845 | |||||
| Recessive mode | 1.723 (1.210-2.453) | 0.003 | 0.831 (0.500-1.380) | 0.474 | |||||
| Additive model | 0.894 (0.732-1.092) | 0.273 | 1.037 (0.767-1.403) | 0.814 | |||||
| KIR3DL2 | rs1654644 | ||||||||
| TT | 711 (53.3) | 295 (51.85) | 168 (52.01) | 463 (51.91) | 1 | 1 | |||
| TG | 537 (40.25) | 220 (38.66) | 127 (39.32) | 347 (38.9) | 0.980 (0.797-1.205) | 0.849 | 0.990 (0.724-1.353) | 0.948 | |
| GG | 86 (6.45) | 54 (9.49) | 28 (8.67) | 82 (9.19) | 1.560 (1.083-2.249) | 0.017 | 0.868 (0.512-1.471) | 0.598 | |
| Dominant model | 1.060 (0.872-1.290) | 0.556 | 0.965 (0.719-1.295) | 0.811 | |||||
| Recessive mode | 1.574 (1.104-2.243) | 0.012 | 0.871 (0.522-1.454) | 0.598 | |||||
| Additive model | 0.924 (0.756-1.129) | 0.439 | 1.011 (0.747-1.368) | 0.944 | |||||
| HLA-A | rs3869062 | ||||||||
| AA | 882 (66.97) | 367 (65.54) | 196 (60.87) | 563 (63.83) | 1 | 1 | |||
| AG | 386 (29.31) | 170 (30.36) | 113 (35.09) | 283 (32.09) | 1.251 (1.010-1.550) | 0.04 | 1.205 (0.877-1.656) | 0.25 | |
| GG | 49 (3.72) | 23 (4.11) | 13 (4.04) | 36 (4.08) | 1.242 (0.752-2.051) | 0.396 | 1.140 (0.546-2.382) | 0.727 | |
| Dominant model | 1.250 (1.017-1.535) | 0.034 | 1.197 (0.882-1.626) | 0.248 | |||||
| Recessive mode | 1.155 (0.703-1.898) | 0.568 | 1.068 (0.516-2.214) | 0.859 | |||||
| Additive model | 1.235 (0.999-1.527) | 0.051 | 1.195 (0.873-1.636) | 0.267 | |||||
| HLA-A | rs12202296 | ||||||||
| TT | 448 (33.48) | 184 (32.34) | 122 (37.77) | 306 (34.3) | 1 | 1 | |||
| TC | 647 (48.36) | 286 (50.26) | 160 (49.54) | 446 (50.0) | 0.992 (0.799-1.232) | 0.944 | 0.924 (0.669-1.277) | 0.631 | |
| CC | 243 (18.16) | 99 (17.4) | 41 (12.69) | 140 (15.7) | 0.789 (0.586-1.063) | 0.12 | 0.694 (0.435-1.107) | 0.125 | |
| Dominant model | 0.938 (0.764-1.152) | 0.542 | 0.867 (0.637-1.181) | 0.365 | |||||
| Recessive mode | 0.793 (0.606-1.038) | 0.091 | 0.727 (0.475-1.114) | 0.143 | |||||
| Additive model | 1.070 (0.880-1.301) | 0.498 | 1.029 (0.766-1.382) | 0.849 |
- Note: p Value, OR, and 95% CIs of (a)Group (B + C) Ver A where (b)for Group C Ver B, were computed on the basis of the logistic regression model, adjusted by sex, age, ALT, AST, IFNL4 rs12979860, IFNL4 rs8099917, and infection route. Bolded text represents substantially significant outcomes.
- Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; Group A, uninfected control group; Group B, persistent infection group; Group C, spontaneous clearance group; HLA, human leukocyte antigen; IFNL4, interferon lambda 4, also know as IL28B; KIR, killer-cell immunoglobulin-like receptors; SNPs, single nucleotide polymorphisms.
The four significant SNPs before multiple corrections in Table 2 were included for further exploratory stratified analysis to control for confounding variables, including gender, age, ALT, AST, and routes of infection, which may introduce bias. Compared with the wild allele carriage of KIR3DL2 rs11672983, KIR3DL2 rs3745902, KIR3DL2 rs1654644, and HLA-A rs3869062, a significant higher risk was found in the mutant allele carriage of these SNPs in some subgroups (all p < 0.05, showed in Table 3).
| Gene | SNPs | Subgroups | Group A | Group (B + C) | OR (95%CI)a | pa |
|---|---|---|---|---|---|---|
| n (WW/WM/MM) | n (WW/WM/MM) | |||||
| KIR3DL2 | rs11672983 | Age | ||||
| <50 | 184/115/16 | 108/80/19 | 1.650 (0.742-3.671) | 0.22 | ||
| ≥50 | 538/414/70 | 359/264/65 | 1.555 (1.051-2.300) | 0.027 | ||
| Gender | ||||||
| Male | 266/213/30 | 141/88/29 | 1.971 (1.065-3.645) | 0.031 | ||
| Female | 456/316/56 | 326/256/55 | 1.494 (0.963-2.318) | 0.074 | ||
| ALT (U/L) | ||||||
| <50 | 702/512/85 | 343/258/61 | 1.591 (1.109-2.281) | 0.012 | ||
| ≥50 | 16/11/1 | 123/84/23 | 3.288 (0.383-28.198) | 0.278 | ||
| AST (U/L) | ||||||
| <40 | 686/501/81 | 304/229/60 | 1.714 (1.194-2.461) | 0.004 | ||
| ≥40 | 32/22/5 | 162/113/24 | 0.692 (0.211-2.271) | 0.543 | ||
| Route of infection | ||||||
| DP | 235/153/26 | 78/59/16 | 1.726 (0.845-3.524) | 0.134 | ||
| PBD | 487/376/60 | 389/285/68 | 1.614 (1.066-2.444) | 0.024 | ||
| KIR3DL2 | rs3745902 | Age | ||||
| <50 | 180/120/16 | 105/83/19 | 1.658 0.745-3.687 | 0.215 | ||
| ≥50 | 526/436/68 | 354/268/66 | 1.662 (1.121-2.463) | 0.011 | ||
| Gender | ||||||
| Male | 262/218/32 | 138/91/29 | 1.878 (1.023-3.447) | 0.042 | ||
| Female | 444/338/52 | 321/260/56 | 1.693 (1.083-2.646) | 0.021 | ||
| ALT (U/L) | ||||||
| <50 | 686/539/83 | 337/262/61 | 1.689 (1.176-2.426) | 0.005 | ||
| ≥50 | 16/11/1 | 121/87/24 | 3.311 (0.386-28.376) | 0.275 | ||
| AST (U/L) | ||||||
| <40 | 671/526/80 | 300/231/60 | 1.767 (1.229-2.540) | 0.002 | ||
| ≥40 | 31/24/4 | 158/118/25 | 1.012 (0.271-3.787) | 0.986 | ||
| Route of infection | ||||||
| DP | 230/158/26 | 76/61/16 | 1.726 (0.845-3.524) | 0.134 | ||
| PBD | 476/398/58 | 383/290/69 | 1.775 (1.167-2.700) | 0.007 | ||
| KIR3DL2 | rs1654644 | Age | ||||
| <50 | 181/118/16 | 108/81/18 | 1.527 (0.675-3.453) | 0.309 | ||
| ≥50 | 530/419/70 | 355/266/64 | 1.530 (1.032-2.269) | 0.034 | ||
| Gender | ||||||
| Male | 261/216/31 | 139/89/29 | 1.911 (1.038-3.518) | 0.038 | ||
| Female | 450/321/55 | 324/258/53 | 1.452 (0.929-2.270) | 0.101 | ||
| ALT (U/L) | ||||||
| <50 | 691/520/85 | 342/260/59 | 1.539 (1.070-2.214) | 0.02 | ||
| ≥50 | 16/11/1 | 120/85/23 | 3.314 (0.387-28.383) | 0.274 | ||
| AST (U/L) | ||||||
| <40 | 676/508/81 | 302/231/58 | 1.660 (1.152-2.390) | 0.007 | ||
| ≥40 | 31/23/5 | 160/114/24 | 0.693(0.211-2.274) | 0.545 | ||
| Route of infection | ||||||
| DP | 232/155/26 | 78/59/16 | 1.730 (0.847-3.530) | 0.132 | ||
| PBD | 479/382/60 | 385/288/66 | 1.558 (1.024-2.370) | 0.038 | ||
| HLA-A | rs3869062 | Age | ||||
| <50 | 214/91/7 | 128/68/9 | 1.585 (1.027-2.446) | 0.038 | ||
| ≥50 | 668/295/42 | 435/215/27 | 1.220 (0.965-1.543) | 0.097 | ||
| Gender | ||||||
| Male | 334/150/18 | 169/74/10 | 1.476 (1.015-2.147) | 0.041 | ||
| Female | 548/236/31 | 394/209/26 | 1.162 (0.903-1.496) | 0.244 | ||
| ALT (U/L) | ||||||
| <50 | 854/377/48 | 403/222/27 | 1.223 (0.990-1.510) | 0.062 | ||
| ≥50 | 23/4/1 | 158/60/9 | 2.026 (0.686-5.988) | 0.201 | ||
| AST (U/L) | ||||||
| <40 | 831/369/48 | 367/192/24 | 1.207 (0.973-1.497) | 0.087 | ||
| ≥40 | 46/12/1 | 194/90/12 | 2.039 (0.932-4.465) | 0.075 | ||
| Route of infection | ||||||
| DP | 276/119/11 | 88/57/6 | 1.715 (1.105-2.663) | 0.016 | ||
| PBD | 606/267/38 | 475/226/30 | 1.139 (0.895-1.449) | 0.289 |
- Note: Bold type indicates statistically significant results.
- Abbreviations: HCV, hepatitis C virus; OR, odds ratio; CI, confidence interval; DP, dialysis population; Group A, uninfected control group; Group B, persistent infection group; Group C, spontaneous clearance group; Group (B + C): infected group; PBD, paid blood donors; M, mutant-type allele; W, wild-type allele.
- a p Value, OR and 95% CIs of Group A versus Group (B + C) were calculated based on the logistic regression model, adjusted by gender, age, ALT, AST, IFNL4 rs12979860, IFNL4 rs8099917 and route of infection;
3.3 Association of candidate SNPs with the spontaneous viral clearance of HCV infection
The relationship between the candidate SNPs and the spontaneous viral clearance of HCV infection was analyzed by comparing the SNP distribution frequencies between Group B (individuals with persistent infection) and Group C (patients who achieved spontaneous viral clearance). However, no association was found between the five candidate SNPs (KIR3DL2 rs11672983, KIR3DL2 rs3745902, KIR3DL2 rs1654644, HLA-A rs3869062 and HLA-A rs12202296) and the spontaneous clearance of HCV in the logistic regression analysis using four genetic models, adjusting for gender, age, ALT, AST, IFNL4 rs12979860 and IFNL4 rs8099917, the route of infection (all p > 0.05, Table 2). Therefore, no further stratified analysis was performed on these data. Due to the high amount of missing HCV genotype data, it was difficult to include in the logistic regression analysis. To obtain more rigorous results, we conducted a further analysis on patients with complete HCV genotype data. The results indicate that the conclusion remains unchanged regardless of whether the HCV genotype is included as a covariate (data no shown).
3.4 Combined effects analysis
The combined effects of positive SNPs, KIR3DL2 rs11672983 and KIR3DL2 rs3745902, on the susceptibility to HCV infection was assessed by counting the number of their risk genotypes (rs11672983 AA and rs3745902 TT). The risk of the susceptibility to HCV infection increased with the presence of more unfavorable genotypes (pTrend = 0.007, Table 4), and carrying rs11672983 AA or rs3745902 TT genotypes was more susceptible to HCV infection (adjusted OR = 1.628, 95% CI = 1.151−2.302, p = 0.006), while carrying all two unfavorable genotypes correlated to the highest risk (adjusted OR = 1.711, 95% CI = 1.194−2.450, p = 0.003). The results of the combined effects analysis were adjusted for confounding factors through logistic regression.
| Risk genotypesa | Group A n (%) |
Group (B + C) n (%) |
HCV-infection rate (%) |
OR (95% CI)b | pb |
|---|---|---|---|---|---|
| 0 | 1244 (93.32) | 806 (90.36) | 39.32 | 1 | |
| 1 | 8 (0.6) | 4 (0.45) | 33.33 | 0.880 (0.248-3.116) | 0.843 |
| 2 | 81 (6.08) | 82 (9.19) | 50.31 | 1.711 (1.194-2.450) | 0.003 |
| Trend | 0.007c | ||||
| 0 | 1244 (93.32) | 806 (90.36) | 39.32 | 1 | |
| 1-2 | 89 (6.68) | 86 (9.64) | 49.14 | 1.628 (1.151-2.302) | 0.006 |
- Note: Bold type indicates statistically significant results.
- Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; CI, confidence interval; Group A: uninfected control group; Group B: persistent infection group; Group C: spontaneous clearance group; Group (B + C): infected group; OR, odds ratio.
- a Number of unfavorable genotypes (rs11672983-AA and rs3745902-TT);
- b p Value, OR, and 95% CIs of Group A and Group (B + C) were computed according to the logistic regression model, adjusted by sex, age, ALT, AST, IFNL4 rs12979860, IFNL4 rs8099917, and route of infection;
- c p Value for the Linear by Linear Test.
3.5 Bioinformatics analysis
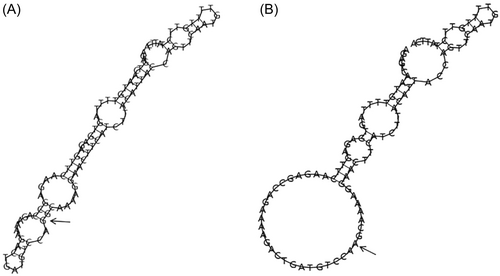
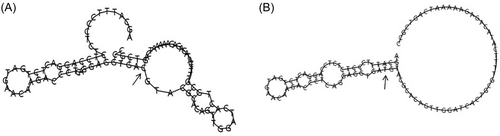
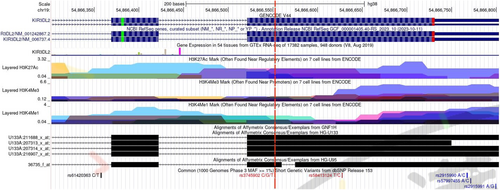
The SNPinfo Web server predicts that KIR3DL2 rs11672983 is predicted to serve as a transcription factor binding sites (TFBS). Meanwhile, KIR3DL2 rs3745902 may function as exonic splicing enhancers or silencers (ESE/ESS) and nonsynonymous SNPs (nsSNPs). The RegulomeDB scores for the two SNPs, rs11672983 and rs3745902, are both “1f,” indicating potential involvement in expression quantitative trait loci (eQTL) or chromatin accessibility quantitative trait loci (caQTL). These loci could influence gene expression or chromatin accessibility in specific regions, such as TFBS or chromatin open peaks. Furthermore, the impact of mutations at the rs11672983 and rs3745902 loci on the secondary structure of KIR3DL2 mRNA was analyzed using the RNAfold web server. The results, depicted in Figure 1 and 2, reveal significant differences between the wild type and mutant type of rs11672983 and rs3745902, indicating alterations in the KIR3DL2 mRNA secondary structure. In addition, the potential biological functions of the two SNPs were annotated in the ENCODE project using the UCSC genome browser. rs3745902 was found to be located on elevated peaks of H3K4Me3 and H3K27Ac markers in seven cell lines. This positioning was confirmed by the enrichment of H3K4Me3 and H3K27Ac observed in CHIP-seq assays (Figure 3).
4 DISCUSSION
In this study, we investigated the role of genetic variants in the KIR3DL2 and HLA-A genes in HCV infection, including five candidate SNPs: KIR3DL2 rs11672983, KIR3DL2 rs3745902, KIR3DL2 rs1654644, HLA-A rs3869062, and HLA-A rs12202296. The data indicate that KIR3DL2 rs11672983 AA and KIR3DL2 rs3745902 TT are associated with an increased susceptibility to HCV infection. Stratified analysis further confirmed the association between the carriage of these risk genotypes and the susceptibility to HCV infection in some subgroups. In addition, the risk of the susceptibility to HCV infection increased with the presence of more unfavorable genotypes. Last but not least, the results of bioinformatics analysis imply that the variations at rs11672983 and rs3745902 may play a regulatory role in transcriptional and translational mechanisms. The two distinctive features of this study are, firstly, the large sample size, and secondly, the adjustment of all results for confounding factors, including IFNL4 rs12979860 and IFNL4 rs8099917, both of which have been validated to possess a significant association with HCV susceptibility, spontaneous viral clearance and the therapeutic efficacy of the peg-IFN-α and ribavirin regimen.28, 29
KIR3DL2 and HLA-A were chosen based on the intricate receptor-ligand interactions of KIR/HLA and their functional significance in HCV immune responses. The promoter region of KIR3DL2 stands out within the KIR family promoters, featuring multiple potential nuclear factor-kB binding sites.36 Positioned as an inhibitory receptor in the intricately functional KIR gene cluster B type, KIR3DL2 frequently exerts inhibitory effects on NK cells and T lymphocytes, contributing to the regulation of the immune system's functions.37 In a recent study, Decroos et al. proposed KIR3DL2 as a promising candidate for enhancing the management of aggressive systemic peripheral T-cell lymphomas.38 Additionally, KIR3DL2 binds to multiple HLA-A ligands. HLA-A has also been demonstrated to be associated with varying degrees of liver fibrosis and hepatocellular carcinoma related to HCV.39, 40 Therefore, KIR3DL2 and HLA-A were chosen for investigation in this study, and subsequently, a specific screening strategy was employed to select candidate SNPs.
In logistic regression analysis, we found that the rs11672983-A and rs3745902-T of KIR3DL2 are associated with an increased risk of HCV. KIR3DL2 rs11672983 is located within the 3' UTR region of the KIR3DL2 gene. Genetic variations in noncoding regions impact susceptibility to infections in various ways. For example, SNPs located in promoter and enhancer regions can influence the binding of transcription factors, thereby regulating gene expression.41 The splicing process of mRNA can be affected by sequence variations in noncoding regions, leading to changes in splicing patterns. This may result in the production of different protein isoforms, or affect the stability and translational efficiency of mRNA, thereby impacting protein function and the host's response to infection. Additionally, SNPs in the 3′UTRs may affect the binding of miRNAs, which regulate the stability and translation of mRNA through their interaction. Furthermore, the three-dimensional structure of the genome can regulate gene expression by promoting spatial proximity and interactions between distant sites.42 These elements collectively determine the transcription and subsequent processing of the gene. Predictive results from SNPinfo suggest that rs3745902 may act as a regulatory element, such as an ESE/ESS or nsSNP, involved in the expression of the KIR3DL2 gene. Another positive SNP, rs3745902, is located in exon 9 of KIR3DL2 and represents a missense mutation. The allelic change from C to T at this locus results in the substitution of the 11th amino acid in the protein from serine to methionine, potentially impacting protein expression. Chromatin analysis using the UCSC Genome Browser revealed that rs3745902 is located at the peaks of H3K4Me1 and H3K27Ac histone modifications, both of which are markers of active regulatory sequences within chromatin domains.43 Since aberrant histone modifications can precipitate deviant gene expression patterns, they may be instrumental in the pathogenesis of inflammatory disorders and a spectrum of neoplasms. Notably, H3K4Me3 and H3K27Ac are predominantly accumulated at loci of vigorous gene transcription, ostensibly facilitating and amplifying this cardinal biological process.44 H3K4me3 has been correlated with the forecasting of clinical outcomes in a subset of malignancies, including hepatic and cervical carcinomas.45, 46 Analysis of eQTL confirmed the impact of genetic variations at rs11672983 and rs3745902 on gene expression levels, with the mutant A allele of rs11672983 and the mutant T allele of rs3745902 associated with enhanced expression levels. Considering that KIR3DL2 is an inhibitory receptor on the surface of NK cells, it is speculated that the upregulation of KIR3DL2 expression caused by these mutations may be associated with the inhibition of NK cell function. Therefore, these SNP variations may enhance the inhibitory capacity of NK cells by upregulating KIR3DL2, thereby influencing innate immune mechanisms and regulating susceptibility to HCV infection.
Based on age, gender, ALT, AST, and high-risk population, stratified analysis of KIR3DL2 rs11672983 and rs3745902 revealed that the rs11672983-A and rs3745902-T alleles also increase susceptibility to HCV in certain subgroups. Research has historically underscored the pivotal roles of both age and sex in shaping the trajectory of HCV infections, with youth and female gender seemingly offering a degree of protection. Such protective mechanisms could stem from differential immune responses associated with age, as well as the regulatory influences that estradiol and estrogen receptors exert on the body's innate and acquired immunity systems.47, 48 Additionally, considering the potential for different routes of infection to result in varying HCV inoculum sizes and immune response intensities, the rs11672983-A allele and rs3745902-T allele are associated with increased susceptibility among repeat compensated blood donors. All these findings indicate a complex interplay among age, gender, high-risk populations, and genetic factors. Although KIR3DL2 rs1654644 and HLA-A rs3869062 were weakly associated with HCV susceptibility and subsequently excluded after multiple corrections, stratified analysis suggests that these loci are also potential contributors to increased HCV susceptibility.
In this study, there are several additional questions. Firstly, the participants involved in this study had engaged in paid blood donation before the implementation of the Blood Donation Law in China in 1998. Secondly, obtaining accurate information from participants regarding the timing of their initial infection and the volume of blood transfusions they received poses a challenge. Thirdly, to minimize selection bias, these three groups were drawn from the same population. Moreover, we adjusted for various confounding factors, including age, gender, ALT, AST, route of infection, through logistic regression. Particularly, we also included the genotypes of IFNL4 rs12979860 and IFNL4 rs8099917 in the adjustment model. Lastly, further genetic and functional studies are needed to determine the mechanisms of KIR3DL2/HLA-A genetic variations in the process of HCV infection.
5 CONCLUSION
In summary, the findings of this study indicate a significant association between KIR3DL2/HLA-A gene variations and increased susceptibility to HCV infection. These genetic markers may modulate the innate immune response by influencing the expression patterns of KIR3DL2/HLA-A in the context of HCV infection.
AUTHOR CONTRIBUTIONS
Ming Yue and Peng Huang designed the research; Yuwen Li, Tian Zeng, Zepei Feng, Chao Shen, Haozhi Fan, Wen Yin, Liqin Qian and Chengrui Ren performed the research; Tian Zeng, Ming Yue and Peng Huang analyzed data and wrote the article; Weilong Tan, and Haozhi Fan checked the statistical calculations; Yue Feng, Xueshan Xia and Chuanlong Zhu commented on and revised the paper. All authors have read and approved the final paper.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (82273691), Key Project of Natural Science Foundation of Yunnan Province (2019FA005), the Science and Technology Plan of Hainan Province (Clinical Research Center) (LCYX202204 and LCYX202306), the Hainan Province Science and Technology Special Fund (ZDYF2022SHFZ067), and the grants from Jiangsu Provincial Medical Key Discipline (Laboratory) Cultivation Unit.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict to interest.
ETHICS STATEMENT
The studies involving human participants were reviewed and approved by the Institutional Ethics Review Committee of Nanjing Medical University (Nanjing, China). The patients/participants provided their written informed consent to participate in this study.
Open Research
DATA AVAILABILITY STATEMENT
According to national legislation/guidelines, specifically the Administrative Regulations of the People's Republic of China on Human Genetic Resources (http://www.gov.cn/zhengce/content/2019-06/10/content_5398829.htm, http://english.www.gov.cn/policies/latest_releases/2019/06/10/content_281476708945462.htm), no additional raw data is available at this time. Data of this project can be accessed after an approval application to the China National Genebank (CNGB, https://db.cngb.org/cnsa/). Please refer to https://db.cngb.org/, or email: [email protected] for detailed application guidance. The accession code CNP0001926 should be included in the application.




