Comparable CD8+ T-cell responses to SARS-CoV-2 vaccination in single-cell transcriptomics of recently allogeneic transplanted patients and healthy individuals
Benedikt Obermayer and Il-Kang Na contributed equally to this study.
Abstract
Despite extensive research on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccination responses in healthy individuals, there is comparatively little known beyond antibody titers and T-cell responses in the vulnerable cohort of patients after allogeneic hematopoietic stem cell transplantation (ASCT). In this study, we assessed the serological response and performed longitudinal multimodal analyses including T-cell functionality and single-cell RNA sequencing combined with T cell receptor (TCR)/B cell receptor (BCR) profiling in the context of BNT162b2 vaccination in ASCT patients. In addition, these data were compared to publicly available data sets of healthy vaccinees. Protective antibody titers were achieved in 40% of patients. We identified a distorted B- and T-cell distribution, a reduced TCR diversity, and increased levels of exhaustion marker expression as possible causes for the poorer vaccine response rates in ASCT patients. Immunoglobulin heavy chain gene rearrangement after vaccination proved to be highly variable in ASCT patients. Changes in TCRα and TCRβ gene rearrangement after vaccination differed from patterns observed in healthy vaccinees. Crucially, ASCT patients elicited comparable proportions of SARS-CoV-2 vaccine-induced (VI) CD8+ T-cells, characterized by a distinct gene expression pattern that is associated with SARS-CoV-2 specificity in healthy individuals. Our study underlines the impaired immune system and thus the lower vaccine response rates in ASCT patients. However, since protective vaccine responses and VI CD8+ T-cells can be induced in part of ASCT patients, our data advocate early posttransplant vaccination due to the high risk of infection in this vulnerable group.
1 INTRODUCTION
COVID-19 disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has accounted for significant morbidity and mortality worldwide. The development and approval of several SARS-CoV-2 vaccines has progressed at astonishing speed, and excellent vaccine response and adequate protection against severe disease progression have been demonstrated in healthy individuals.1-3 However, numerous studies have shown that hematologic patients in general, and allogeneic hematopoietic stem cell transplantation (ASCT) patients in particular, have a significantly weaker vaccine response than healthy controls, with a substantial number of patients failing to develop a protective immunity at all.4-14 The current knowledge of vaccine response in patients after ASCT is limited to data on measurements of serological responses and in vitro measurements of specific T-cell immunity at various, commonly late time points after transplantation.
Apart from ASCT patients, single-cell RNA sequencing (scRNA-seq) and bulk sequencing strategies have been applied to dissect immune responses to SARS-CoV-2 infection and vaccination, providing in-depth insights into changes in immune cell abundances and gene expression.15-21 Complimented by B cell receptor (BCR) and T cell receptor (TCR) sequencing, analysis of B- and T-cell receptor repertoires and dynamics of specific clonotypes have been performed and a considerable number of SARS-CoV-2-specific TCR and antibody sequences have been described.22-26 Despite a common vaccination antigen, only a fraction of these responses can be attributed to public TCRs and BCRs, whereas a large proportion of the SARS-CoV-2-specific response is accounted for by private clonotypes. In a recent study, Zhang et al. described a subgroup of CD8+ T-cells exhibiting a characteristic gene expression profile highly associated with SARS-CoV-2 specificity.27 The authors further observed a correlation between the abundance of this cell group and the severity of infection, suggesting an important role in combating SARS-CoV-2 infection.
In this study, we augmented SARS-CoV-2-specific serology and T-cell functionality with scRNA-seq and BCR/TCR profiling for the first time in ASCT patients, which allowed us to investigate potential molecular parameters for the weaker vaccine responses in ASCT patients and to characterize vaccine-induced (VI) T- and B-cell clonotypes. Further, we compared our ASCT scRNA-seq data with that of healthy vaccinees from open data repositories utilizing label transfer from a recently published reference,27 to evaluate and predict vaccination-mediated immunity according to expression signatures in the adaptive immune response of ASCT patients.
2 MATERIALS AND METHODS
2.1 Patients
Peripheral blood from ASCT recipients was collected before the first (T0), after the first (T1) and after the second vaccine shot (T2) with BNT162b2 during June 2021–August 2022. Patient characteristics are summarized in Table 1. None of the patients had a history of SARS-CoV-2 infection before immunization. On average, the first vaccination was administered on day 146 (±44) after ASCT, eight out of 15 patients were free of immunosuppressive therapy at the time of sample collection and initiation of vaccination course. Three patients (ASCT1–3) were analyzed in-depth via paired scRNA-seq and TCR/BCR profiling. ASCT3 suffered disease relapse around day +30 after ASCT and was successfully treated with gilteritinib and donor lymphocyte infusions and remained in remission throughout our sampling period. The same patient experienced a breakthrough infection around 4 months after second vaccination and was treated with casirivimab/imdevimab. ASCT2 experienced disease relapse around 90 days after the second vaccine shot and eventually succumbed to his disease, hence was not available for later analyses.
| Days from Tx | Days from 1st vaccination to evaluation | Days from 2nd vaccination to evaluation | Response | Age | Sex | Diagnosis | Conditioning | Donor type | IS | ATG | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASCT 1 | 162 | 12 | 16 | Yes | 33 | F | AML | MAC | MUD | No | Yes |
| ASCT 2 | 185 | 12 | 15 | Yes | 36 | M | AML | RIC | MUD | No | Yes |
| ASCT 3 | 199 | 18 | 17 | No | 67 | M | AML | MAC | MRD | No | Yes |
| ASCT 4 | 195 | n/a | 71 | No | 62 | M | MPN | RIC | MUD | No | Yes |
| ASCT 5 | 196 | n/a | 41 | No | 62 | M | AML | RIC | MMUD | Yes | Yes |
| ASCT 6 | 146 | n/a | 16 | No | 59 | M | MDS | RIC | MUD | No | Yes |
| ASCT 7 | 40 | n/a | 117 | No | 67 | M | MDS | RIC | MUD | Yes | Yes |
| ASCT 8 | 120 | n/a | 129 | No | 70 | M | AML | RIC | MUD | No | Yes |
| ASCT 9 | 107 | n/a | 119 | No | 46 | M | NHL | RIC | MUD | Yes | Yes |
| ASCT 10 | 129 | n/a | 35 | No | 74 | M | NHL | RIC | MUD | Yes | Yes |
| ASCT 11 | 200 | n/a | 26 | Yes | 59 | M | MDS | RIC | MRD | Yes | Yes |
| ASCT 12 | 138 | n/a | 10 | Yes | 52 | M | AML | MAC | MRD | Yes | No |
| ASCT 13 | 120 | n/a | 146 | Yes | 68 | M | AML | RIC | MMUD | No | Yes |
| ASCT 14 | 123 | n/a | 59 | Yes | 73 | F | AML | RIC | MMUD | Yes | Yes |
| ASCT 15 | 152 | n/a | 152 | Yes | 58 | M | AML | RIC | MUD | No | Yes |
| ASCT 16a | 189 | n/a | n/a | n/a | 53 | F | MCL | MAC | MRD | n/a | Yes |
| ASCT 17a | 178 | n/a | n/a | n/a | 56 | F | BDPCN | MAC | Haplo | n/a | No |
| ASCT 18a | 180 | n/a | n/a | n/a | 65 | M | AML | MAC | MRD | n/a | Yes |
| ASCT 19a | 179 | n/a | n/a | n/a | 54 | M | MDS | MAC | MRD | n/a | Yes |
- Abbreviations: AML, acute myeloid leukemia; ASCT, allogeneic hematopoietic stem cell transplantation; ATG, antithymocyte globulin; BDPCN, blastic plasmacytoid dendritic cell neoplasm; haplo, haploidentical transplantation; IS, medicinal immunosuppression at the time of vaccination; MAC, myeloablative conditioning; MCL, mantle cell lymphoma; MDS, myelodysplastic syndrome; MMUD, mismatched unrelated donor; MPN, myeloproliferative disease; MRD, matched related donor; MUD, matched unrelated donor; NHL, non-Hodgkin lymphoma; RIC, reduced intensity conditioning; Tx, Transplantation.
- a Unvaccinated ASCT recipients from Obermayer et al.28
2.2 Peripheral mononuclear blood cell (PBMC) preparation
PBMCs were isolated from heparinized whole blood by density gradient centrifugation and frozen at −80°C until further analysis.
2.3 Serology
Antibody titers against antispike IgG and IgA antibody titers against SARS-CoV-2 spike protein were measured after the second vaccine dose using Elecsys Anti-SARS-CoV-2 S; Roche Diagnostics.
2.4 Ex vivo T-cell stimulation
Fresh or frozen PBMCs were cultivated at a concentration of 5 × 106 cells in RPMI 1640 medium (Gibco) supplemented with 10% heat-inactivated AB serum (Pan Biotech), 100 U/mL penicillin (Biochrom), 0.1 mg/mL streptomycin (Biochrom). Stimulations were performed with PepMixTM overlapping peptide pools (15-aa length with 11-aa overlaps; JPT Peptide Technologies) including the N-terminal (S-I) or C-terminal (S-II) part of spike glycoprotein, NCAP-1 (N), VME-1 (M), AP3A (ORF3a), NS7A, NS8, Pan-SARS2select-1 (dominant epitopes including spike), Pan-SARS2select-2 (dominant epitopes without spike) and custom-made pools containing cross-reactive epitopes with spike or without spike and the single peptide S816-830 (N′-SFIEDLLFNKVTLAD-C′). All stimulations were conducted at concentrations of 1 µg/mL per peptide. Equal concentrations of dimethyl sulfoxide in phosphate-buffered saline as negative control and CEFX Ultra SuperStim pool (JPT Peptide Technologies) as positive control were used. Incubation was performed at 37°C, 5% CO2 for 16 h with 10 μg/mL Brefeldin A (Sigma-Aldrich) added 2 h after starting the stimulation.
2.5 Flow cytometry
After stimulation, staining of surface antigens and intracellular staining were carried out with fluorochrome-conjugated antibodies titrated to their optimal concentrations (Tables S1 and S2). To exclude dead cells, Zombie Yellow fixable viability staining (BioLegend) was added. Fixation and permeabilization were performed with eBioscience FoxP3 fixation and PermBuffer (Invitrogen) according to the manufacturer's protocol. Samples were measured on a MACSQuant Analyzer 16 using MACSQuantify software (v.2.13). Instrument performance was monitored daily with Rainbow Calibration Particles (BD). Antigen-specific T-cell responses were determined based on 4-1BB and CD40L expression in CD4+ T-cells and IFNγ and TNF-α expression in CD8+ T-cells. For gating strategy see Figure S1A.
2.6 5′ scRNA-seq
After thawing of PBMCs, we treated samples individually, using one 10× lane per sample. Samples were converted to barcoded scRNA-seq libraries using a Chromium Next GEM Single Cell V(D)J Reagent Kit (10x Genomics; Pleasanton) according to manufacturer's instructions. 5′ Library Kit was used to prepare single-cell RNA libraries according to the manufacturer's protocol. Full-length TCR/BCR V(D)J segments were enriched from amplified cDNA from 5′ libraries via PCR amplification using a Chromium Single-Cell V(D)J Enrichment Kit according to the manufacturer's protocol (10x Genomics). The mRNA library average sequencing depth aimed for was 10 000 read pairs per cell and 5000 read pairs per cell for the V(D)J libraries. Sequencing of the resulting libraries was performed on an Illumina NovaSeqxx.
2.7 Utilized public data
Public single-cell data of healthy controls receiving mRNA SARS-CoV-2 vaccinations were obtained from Sureshchandra et al.18 and Wang et al.16 Raw data of Sureshchandra et al. was downloaded from SRA (accession number PRJNA767017) and processed with cellranger multi (v6.0.0) against the GRCh38 genome annotation and TotalSeq C hashtag barcodes. Pooled samples were demultiplexed using a combination of HTOdemux29 and vireo (v0.5.6)30 after scoring common variants from the 1000Genomes project with cellsnp-lite (v1.2.0).31 Processed data for Wang et al. was downloaded from OMIX (accession number OMIX001295-01).16 We additionally included single-cell data of unvaccinated healthy donors and ASCT recipients from Obermayer et al.,28 which was downloaded from gene expression omnibus (GEO) (accession number GSE222633), and matching BCR data existing in-house but not used in the original study was added. All public data sets were processed with Seurat and scRepertoire as described below. The reference COVID-19 vaccination data set27 was downloaded from Zenodo (https://zenodo.org/records/7555405).
2.8 Bioinformatic analysis and statistics
Sequencing libraries for gene expression and TCR/BCR were jointly processed using cellranger multi (v6.0.0) and the GRCh38 genome annotation. For a consistent cell type annotation across the different data sets and to leverage the increased power of multimodal single-cell data sets to resolve cell type identities, we used Seurat v4.0.132 to jointly transfer cell type labels and embedding coordinates from a recent COVID-19 vaccination scRNA-seq data set,27 which profiled expression of surface proteins along with RNA. This reference mapping workflow based on supervised principal component analysis (PCA) has been shown to improve accuracy and performance of cell type annotation compared to an unsupervised approach.32 We then filtered out cells with more than 10% mitochondrial gene content, less than 250 or more than 5000 genes, those with a level 1 cell type prediction score of less than 0.75, and doublets called by DoubletFinder.33 We used level 2 annotation for B- and T-cells, and level 1 annotation otherwise, and combined the “group_A” and “group_B” VI CD8+ T-cell subtypes. Next, we used scRepertoire v1.1.234 to process cellranger variable, diversifying, and joining segments of the immunoglobulin heavy chain (VDJ) output. VI TCR clonotypes (both chains) were defined as those appearing in at least two time points of each patient. Clonal diversity was assessed using the inverse Simpson score. Antigen specificity was assessed using VDJmatch (v1.3.1) and the VDJdb data base (https://vdjdb.cdr3.net/search).35 Differences in the proportions of VI CD8+ T-cells were tested using mixed-effects binomial models (lme4 package, v1.1-27.1). Isotypes and T cell receptor beta variable (TRBV) and immunoglobulin heavy chain (IGHV) genes were extracted from the cellranger clonotype annotation. Differential usage of TRBV and IGHV genes was tested with binomial models by comparing the fractions of cells using a particular TRBV or IGHV gene across stages. Differential gene expression analysis was performed with DESeq. 2 v1.30.136 using a pseudo-bulk strategy, that is, by summing up counts in all cells of the same type from the same sample. For the comparison of ASCT versus healthy donors prevaccination, we included all control data sets (ASCT, allogeneic stem cell transplantation, controls [ASCT_C], healthy donors 1 [HD1], HD2, HD_C), while the comparison between pre- and postvaccination samples was performed within data sets separately (ASCT, HD1, HD2). Functional enrichment analysis was done with tmod v0.46.237 with gene sets from the Hallmark, Reactome, Kegg, and Gene Ontology BP databases. p Values from differential expression and gene set enrichment analyses were adjusted for multiple comparisons using the Benjamini–Hochberg method. Differences pre- versus postvaccination of the VI GEM scores in CD8+ T-cells were assessed using a linear model with donor identity as covariate.
3 RESULTS
3.1 Humoral and cellular vaccine responses in ASCT patients
Humoral responses to BNT162b2 were longitudinally characterized in 15 patients around 144 days (±46) after ASCT (Figure 1, Table 1, ASCT1–15). Six of these 15 ASCT patients (40%) elicited an antispike-specific IgG response after the second vaccination (Figure 1B). Absolute numbers of CD4+ T-or CD19+ B-cells before vaccination did not correlate with the humoral response (Figure 1C). For a comprehensive assessment of the immune status, we evaluated the SARS-CoV-2-specific T-cell responses and performed scRNA-seq combined with VDJ immunoprofiling before (T0), after the first (T1), and after the second vaccine shot (T2) (Figure 1D). Three patients were selected, each with blood samples available approximately 2 weeks after each immunization. SARS-CoV-2-specific T-cellular responses were assessed by flow cytometry after peptide stimulation (Figure S1). None of the patients showed a SARS-CoV-2-specific T-cell response before vaccination. After vaccinations, ASCT1 and ASCT2 showed a protective CD4+ SARS-CoV-2-specific T-cell response, with that of one patient being significantly stronger (ASCT1). Both these patients also exhibited humoral anti-SARS-CoV-2 responses (Figure 1B). The third patient did not develop a CD4+ T-cell or a humoral SARS-CoV-2-specific immunity (ASCT3). None of the patients developed a significant CD8+ T-cell response after vaccination (Figure S1C). This was also true when using a customized peptide pool with shorter peptides, which are more reliably processed by major histocompatibility complex I molecules (Figure S2A). Around 1 year after the second vaccination, ASCT1 showed a sustained SARS-Cov-2-specific CD4+ T-cell and humoral response; in contrast, ASCT3 did not even show a SARS-CoV-2-specific response after having experienced SARS-CoV-2 “breakthrough infection” in the interim (Figure S2). ASCT2 was not available for this later analysis due to disease relapse.
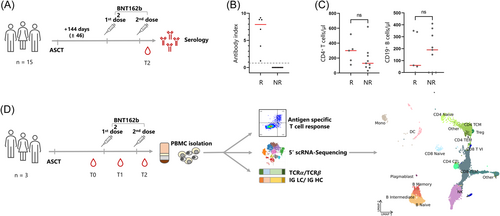
3.2 Differences between ASCT patients and healthy individuals before vaccination
To compare our ASCT data set to data from healthy vaccinees or unvaccinated ASCT patients, we used published single-cell immune profiling data from Sureshchandra et al.18 (in the following HD1), Wang et al.16 (in the following HD2), and Obermayer et al.28 (in the following ASCT_C for the ASCT patients, and HD_C for the associated healthy donors). For characteristics of these healthy donors, see Table 2.
| Donor | Days from 1st vaccination to evaluation | Days from 2nd vaccination to evaluation | Response | Age | Sex |
|---|---|---|---|---|---|
| HD1-D1 | n/a | 14 | Yes | 39 | F |
| HD1-D2 | n/a | 14 | Yes | 45 | F |
| HD1-D3 | n/a | 14 | Yes | 42 | M |
| HD1-D4 | n/a | 14 | Yes | 72 | M |
| HD2-D1 | 21 | 14 | Yes | 35 | M |
| HD2-D2 | 21 | 14 | Yes | 37 | M |
| HD2-D3 | 21 | 14 | Yes | 29 | M |
| HD2-D4 | 21 | 14 | Yes | 31 | F |
| HD2-D5 | 21 | 14 | Yes | 43 | F |
| HD2-D6 | 21 | 14 | Yes | 30 | F |
| HD_C-D1 | n/a | n/a | n/a | 50 | F |
| HD_C-D2 | n/a | n/a | n/a | 23 | F |
| HD_C-D3 | n/a | n/a | n/a | 59 | F |
| HD_C-D4 | n/a | n/a | n/a | 66 | M |
At first, frequencies of immune cell subsets as assessed by scRNA-seq were compared between ASCT patients and published healthy donors before vaccination (Figure 2A). Despite comparable total T-cell numbers, the T-cell compartment of ASCT patients was characterized by a highly altered CD4+/CD8+ ratio in favor of CD8+ T-cells, which accounted for well over 70% of all sequenced cells (Figure 2A). With respect to the B-cell compartment, we noted that ASCT1, ASCT2, ASCT19, and the HD cohorts comprised comparable absolute B-cell numbers. On the other hand, ASCT3, ASCT16, ASCT17, and ASCT18 showed low peripheral B-cell numbers compared to healthy controls. Among ASCT vaccinees, the nonresponder ASCT3 was characterized by very low B-cell numbers (<5% of all sequenced cells) at all investigated time points (Figures 2A and 3A). We next assessed TCR and BCR diversity before vaccination by calculating the inverse Simpson index. As to be expected, TCR diversity was significantly lower in all ASCT patients, when compared to the HD data sets (Figure 2B, left). Analyzing BCR diversity, we found comparable diversity between ASCT patients and HD data sets (Figure 2B, right).
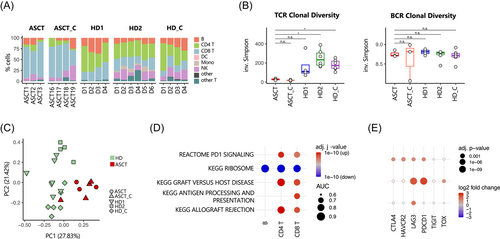

We next quantified compositional differences between data sets using a PCA on the cell type frequencies for each sample and observed a systematic shift between ASCT and HD data sets (Figures 2C and S3A). The systematic differences between ASCT and HD are also reflected on the transcriptome level within different cell types: additional PCAs on gene expression patterns of major immune compartments show a clear separation of ASCT patients and HD samples especially for T-cells (Figure S3B). This separation is driven by a marked upregulation of genes involved in alloreactive activity (Figure 2D). In addition, an exhaustion pattern was evident in CD4+ and CD8+ T-cells as well as in B-cells of ASCT patients with upregulation of LAG3 and programmed cell death protein 1 (PDCD1) (Figure 2E).
Since many factors beyond disease status, from donor characteristics to sample preparation strategies, could lead to systematic batch effects between these data sets, we avoided an unsupervised clustering analysis and cell type discovery and instead used supervised label transfer from a recent COVID-19 vaccination multimodal PBMC scRNA-seq reference data set27 to obtain a consistent high-resolution cell type annotation of all data sets. Indeed, we observed variation between the three HD data sets beyond the clear differences between ASCT and HD (Figures 2A, 3C, and S3A). For instance, HD2 shows a high percentage of naïve CD8+ T-cells,16 while this cell subset is almost completely lacking in the HD1 data18 (Figure 4A). Other cell subsets, such as monocytes and natural killer T cells, are lacking in the HD1 data set probably due to preprocessing effects such as cell sorting. Apart from this, HD1, HD2, and HD_C have a similar distribution of cell subsets (Figure 2A). In addition, all HD data sets share a normal CD4+/CD8+ ratio with an adequate number of CD4+ T-cells.
3.3 Vaccine-related changes in the B-cell compartment of ASCT
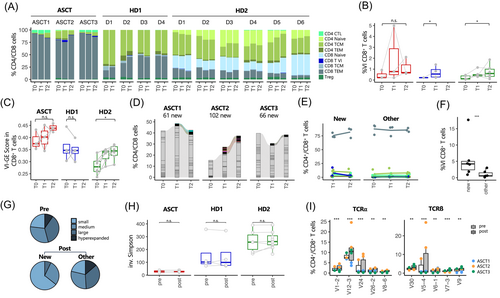
When dissecting B-cell subsets, we found that ASCT1 and ASCT2, the vaccination responders, overwhelmingly possess naïve B-cells, reflecting the naïve repertoire during B-cell reconstitution posttransplant (Figure 3A, time point T0 for ASCT1–3). Nonresponder ASCT3 displayed a more balanced distribution of B-cell subsets; however, ASCT3's absolute B-cell count was significantly lower at all time points. We observed comparable BCR diversity before and after vaccination in our data set, similar to the HD data sets (Figure 3B). Further, there were no significant differences in gene expression detectable when comparing ASCT B-cells before and after vaccination (Figure S4A). Clonal tracking showed that new BCR clonotypes were detectable after the first vaccination (T1) and persisted past the second vaccination (T2) (Figure 3C). These emerging BCR clonotypes presented predominantly with a naïve phenotype; however, a rise in memory B-cell clones was noted (Figure 3D). BCR clones were mostly unique in ASCT patients (Figure 3E). No significant upregulation of specific IGHV genes was evident upon vaccination in ASCT patients; the top five upregulated IGHV genes are depicted in Figure 3F.
3.4 Vaccine-related changes in the T-cell compartment of ASCT
We first analyzed the T-cell subpopulation distribution during vaccinations. No concomitant changes in major T-cell subpopulations were detected in ASCT responders (Figure 4A). Next, we inspected the results of the cell type label transfer from the used reference data set of healthy individuals before and after SARS-CoV-2 vaccination27 and focused on the specific group of VI CD8+ effector memory T-cells, which we were able to detect in all ASCT patients and healthy donors of Sureshchandra et al.18 as well as Wang et al.16 These VI CD8+ T-cells were previously associated with SARS-CoV-2 specificity and characterized by a specific gene set (vaccine-induced gene expression [VI-GE], published in Zhang et al.27 and listed in Table S3). Further, the relative frequency of VI CD8+ T-cells has been shown to be predictive of subsequent clinical outcome.27 As shown for the cohort of Zhang et al.,27 the VI CD8+ T-cell population is present to a small extent even before vaccination (T0), but increases after vaccinations (T1 and T2, Figure 4B). Analyzing the mean expression of the VI-GE profile in single cells, we found an induction of this gene set due to vaccination in ASCT and HD2 data sets, although this only reached statistical significance in HD2 (Figure 4C). In the next step, TCR clonotypes emerging after the first vaccination (T1) and persisting past the second vaccination (T2) were tracked, in the following called newly emerging clonotypes (Figure 4D). Most of the newly emerging TCR clonotypes showed a CD8+ effector memory phenotype and to a much lesser extent a CD4+ central memory phenotype (Figure 4E). Interestingly, we observed a significantly higher fraction of VI CD8+ T-cells within the group of newly emerging clonotypes, suggesting an evolving SARS-CoV-2-specific CD8+ T-cell response (Figure 4F). Sizes of newly emerging clonotypes were highly variable between individuals and comprised mostly of small- and medium-sized clones (Figure 4G). Further, clonal diversity of TCR repertoire and CDR3 length did not change in ASCT patients due to vaccination (Figures 4H and S4B). We observed significant changes in TCRα and TCRβ gene usage after vaccination, with an increased usage of TRAV1-2, TRAV12-3, TRAV24, TRAV26-2, TRAV8-6, TRBV27, TRBV30, TRBV5-4, TRBV7-3, and TRBV9 after vaccination (Figure 4I). Using the VDJmatch and VDJdb databases,35 no SARS-CoV-2 specificity could be assigned to these clonotypes (data not shown). In the comparison of gene profiles of new versus all other clonotypes, we found genes related to an effector memory T-cell phenotype to be induced (GPR183, CD27), whereas genes associated with T-cell dysfunction and senescence were downregulated (FGFBP2, ZF683, ASCL2, PRSS23, Figure S4C). Analyzing differential gene expression of all CD4+ or CD8+ T-cells before and after vaccination, we did not find significant alterations in gene expression of ASCT patients (Figure S4A). Comparing ASCT responders (ASCT1 and ASCT2) with ASCT nonresponder (ASCT3), no significant differences between T-cell subpopulation distribution, gene expression, or TCR gene rearrangements were observed (Figure 4A,D,I).
4 DISCUSSION
The global pandemic has had a tight grip on the world for more than 3 years, but the stranglehold has loosened, due in large part to the rapid development of vaccines. Especially for patients after ASCT, who are at high risk for severe disease progression, knowledge of vaccine efficacy is critical. To understand the detailed effects of the widely used SARS-CoV-2 mRNA vaccine BNT162b on the adaptive immune system of ASCT patients, we examined the serological, cellular, and transcriptomic vaccine responses in ASCT patients and compared them with those of HD.
SARS-CoV-2 mRNA vaccines fail to elicit as sufficient immune responses in ASCT patients as achieved in HD, with seroconversion rates ranging from 50% to 83%6, 8, 10, 12 and SARS-CoV-2-specific T-cell responses in 12%–78%6, 12, 13 of ASCT patients. In our cohort, 40% of ASCT patients presented with seroconversion half year posttransplant. Patient-related factors that have been demonstrated to correlate with poorer vaccine response in ASCT patients are male sex and older age,6 ongoing immunosuppression,8, 10, 12 chronic graft-versus-host disease,12 haploidentical transplant,10 and a short time period (<6 months) between transplantation and vaccination.6, 10, 12 In the presented study, 12 out of 15 patients were transplanted using an HLA-identical donor, around half of the patients had discontinued immunosuppressive therapy before the initiation of vaccination, including ASCT1–3. Concerning the interval between transplantation and vaccination, all investigated patients started vaccination course around 6 months after ASCT (144 ± 46 days). Recommendations for the optimal timing of SARS-CoV-2 vaccination remain controversial. It is known for other vaccinations, for example, influenza and Streptococcus pneumoniae, that responses normalize only 2–3 years after transplantation.38 However, patients who recently underwent ASCT are highly susceptible to infections, so vaccination during this early and vulnerable phase seems clinically appropriate, to protect patients as effectively as possible from potentially life-threatening infections. To this effect, the European Conference on Infections in Leukemia (ECIL) generally recommends to start immunizing with inactivated vaccines as early as 3 months after transplant.38 In accordance with this, the most recent European society for blood and marrow transplantation recommendations state that COVID-19 vaccination in particular shall be initiated as early as 3 months after ASCT, if transmission rates in the surrounding community are high.39 A recently published prospective, multicenter study has provided further evidence of tolerability and noninferiority for vaccination as early as 3 months compared to 4–12 months posttransplant.13
Similar to what has been described before,28 our analysis of differential gene expression revealed significant differences between HD and ASCT patients with a shift toward antigen-driven activation after ASCT particularly in CD4+ T-cells (Figure 2D). In addition to the generally altered gene expression, it was particularly striking that exhaustion markers were upregulated in B- and T-cells from all ASCT patients, which is indicative of the impaired immune system after transplantation (Figure 2E). In accordance with the early time point after transplantation, ASCT patients showed a skewing of the T-cell compartment with marked prominence of CD8+ T-cells and a consequent shift in the CD4+/CD8+ ratio (Figures 2A and 4A). Therefore, it is not surprising that clonally expanded T-cells predominantly exhibit a CD8+ T-cell phenotype (Figure 4E). This opposes the exclusively CD4+ SARS-CoV-2-specific T-cell response we observed in the in vitro assay; here, none of the patients showed a sufficient CD8+ T-cell response (Figure S1C). Similarly contradictory results have been previously reported by Sureshchandra et al.,18 just as it has been previously shown that CD8+ T-cell response after SARS-CoV-2 vaccination is both variable and more feeble.40, 41 Using the VDJmatch and VDJdb databases,35 we were unable to assign SARS-CoV-2 specificity to the new CD8+ T-cell clonotypes detectable after vaccination. However, using label transfer from a previously published reference data set,27 we were able to detect a specific subgroup of VI CD8+ effector memory T-cells in all ASCT patients and healthy donors. This VI CD8+ T-cell subgroup expanded after vaccination and was more pronounced in the group of newly emerging clonotypes, strongly suggesting an evolving SARS-CoV-2-specific CD8+ T-cell response in ASCT patients (Figure 4). Interestingly, our results demonstrate that even the nonresponder ASCT3 exhibits VI CD8+ T-cells, despite mounting neither a serological nor a specific T-cell response in vitro. ASCT3 was the only one of our ASCT patients to suffer an early and symptomatic breakthrough infection approximately 3 months after second vaccination. This may indicate that induction of VI CD8+ T-cells alone is not sufficient to confer protection.
Analyzing overall gene expression in ASCT patients and considering only newly emerging CD8+ T-cell clonotypes that appeared after vaccination and were trackable throughout the second vaccination, we found a significant upregulation in genes associated with an effector memory phenotype as well as a downregulation in genes related to T-cell dysfunction and senescence. While Sureshchandra et al.18 also observed only subtle gene regulations due to vaccinations, Wang et al.16 described a regulation of several thousand genes after vaccination in their donors. In all three studies, time point of sampling after vaccination was identical; however, the type of vaccine differed. In our study and in the work by Sureshchandra et al.,18 the investigated individuals were vaccinated with mRNA vaccines, whereas Wang et al. investigated gene expression after vaccination with an inactivated virus vaccine (CoronaVac). It may be possible that the timing of VI changes in gene expression depends on the type of vaccine used. Corresponding changes after mRNA vaccines could possibly occur off our chosen study time points, and therefore might not have been captured.
In this study, we were not able to identify a preferential IGHV gene usage upon vaccination. In alignment with our results, a study comparing immune cell adaptations after heterologous BNT162b/ChAdOx1 and homologous BNT162b vaccinations in HD found a very diverse induction of immunoglobulin genes after homologous mRNA vaccination, suggesting a highly individual clonal VDJ rearrangement.42 Further underlining the fact that the V-gene repertoire is largely private in nature, other works examining preferential IGHV usage after SARS-CoV-2 vaccination have provided divergent results.16-18, 23 After SARS-CoV-2 infection however, antigen specificity was determined to be a strong driver for IGHV gene usage, with VH1-24, VH3-30, and VH3-33 being the preferentially utilized genes with spike specificity.22-24 Although ASCT responders showed an increase in the usage of the IGHV gene VH3-33 after vaccination, this change did not reach significance (Figure 3F). TCRα and TCRß usage after vaccination was analyzed in ASCT patients and showed TRAV1-2, TRAV12-3, TRAV24, TRAV26-2, TRAV8-6, TRBV27, TRBV30, TRBV5-4, TRBV7-3, and TRBV9 to be preferentially used after vaccination. Interestingly, TRBV27 and TRAV12-3 have been frequently found to be upregulated in convalescent individuals16, 43-47 but have not yet been described after vaccination. In general, TCRα and TCRß utilization in ASCT patients showed high individual variability, highlighting the unique VDJ recombination patterns after contact with the vaccine.
Limitations of our study include the small sample size and the lack of internal controls, resulting in limited statistical power. To leverage additional data from the literature and put our own data in a broader context, we analyzed publicly available data sets of healthy vaccinees as well as of unvaccinated ASCT patients and their associated healthy transplant donors. Our data analysis workflow using cell type label transfer from a common reference data set was designed to make data from different studies comparable and to enhance reproducibility. Despite some remaining batch effects, possibly due to differences in sample (pre)processing, we observe the expected systematic differences between ASCT data and healthy controls (Figure 2). Further comparisons of samples before and after vaccination are performed separately within each data set and should therefore be unaffected by batch effects that may arise from different data sources.
We conclude that ASCT patients can mount a sufficient immune response to SARS-CoV-2 mRNA vaccination within the first 6 months post-ASCT, despite significant distortion of the immune system. When comparing ASCT patients to healthy controls, we found distorted B- and T-cell distribution, reduced TCR diversity, and increased exhaustion marker expression as possible causes for the poorer vaccine response rates in ASCT patients. TCRα/TCRβ and IGHV gene usages were highly variable in ASCT patients. However, we could observe a preferential usage of several TCRα and TCRß chains after SARS-CoV-2 mRNA vaccination. Further, ASCT patients exhibited an increase in CD8+ T-cells after vaccination comprising a gene expression profile predicting SARS-CoV-2 specificity.
Our study establishes a link between certain immune parameters and the known weaker SARS-CoV-2 vaccination response in ASCT patients. Despite expectedly weaker vaccination responses, our results advocate early vaccination after transplantation, as a protective vaccine response and VI CD8+ T-cells can be achieved in ASCT patients despite incomplete immune reconstitution.
AUTHOR CONTRIBUTIONS
Eva Tranter, Benedikt Obermayer and Il-Kang Na conceived and designed the research. Eva Tranter, Marco Frentsch, Benedikt Obermayer and Il-Kang Na interpreted the data and wrote the manuscript. David Busch and Eva Tranter collected and isolated PBMCs from patients. Lucie Loyal and Larissa Henze performed T-cell functionality experimentation. Benedikt Obermayer designed and performed all computational analysis. Marie Luise Hütter-Krönke, Giang Lam Vuong, and Igor-Wolfgang Blau contributed to patient recruitment and clinical management. Andreas Thiel, Lars Bullinger and Dieter Beule commented the manuscript. Stanislav Rosnev participated in finalizing the revised manuscript. All authors edited and approved the final draft of the article.
ACKNOWLEDGMENTS
The authors would like to thank the patients who participated in this study. Luisa Keilholz, a doctorate candidate at Charité Universitätsmedizin Berlin, Germany, supported by the BIH, performed library preparation. Thomas Conrad, senior scientist affiliated with the Core Unit Genomics of the BIH and the Medical Systems Biology at Max Delbrück Center for Molecular Medicine in the Helmholtz Association, Berlin, Germany, carried out the wet lab part of the single-cell sequencing. The project was supported by grants from Berlin Institute of Health at Charité – Universitätsmedizin Berlin, Berlin, Germany (BIH) and research funding from the Stiftung Charité (BIH Johanna Quandt funding). E. T. was a fellowship holder in the BIH-Charité Junior Clinician Scientist Program funded by the Charité – Universitätsmedizin Berlin and the Berlin Institute of Health (BIH).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The study was approved by the Ethics Committee of Charité Universitaetsmedizin Berlin in accordance with the 1964 Declaration of Helsinki and its later amendments (EA1/272/16). All patients gave their written informed consent.
Open Research
DATA AVAILABILITY STATEMENT
Processed data are available from NCBI GEO under accession number GSE253948. All analysis code is available at github.com/bihealth/tranter_vaccination_ASCT.




