Human endogenous retrovirus type K encoded Np9 oncoprotein induces DNA damage response
Jungang Chen and Jiaojiao Fan contributed equally to this study.
Abstract
Human endogenous retrovirus sequences (HERVs) constitute up to 8% of the human genome, yet not all HERVs remain silent passengers within our genomes. Some HERVs, especially the HERV type K (HERV-K), have been found to be frequently transactivated in a variety of inflammatory diseases and human cancers. Np9, a 9-kDa HERV-K encoded protein, has been reported as an oncoprotein and found present in a variety of tumors and transformed cells. In the current study, we for the first time reported that ectopic expression of Np9 protein was able to induce DNA damage response from host cells especially through upregulation of γH2AX. Furthermore, we found that direct knockdown of Np9 by RNAi in Kaposi's Sarcoma-associated herpesvirus (KSHV) infected cells effectively reduced LANA expression, the viral major latent oncoprotein in vitro and in vivo, which may represent a novel strategy against virus-associated malignancies.
1 INTRODUCTION
Human endogenous retrovirus sequences (HERVs) constitute up to 8% of the human genome, and have resided in the human genome for several million years.1, 2 Although the majority of HERVs are dysfunctional due to the accumulation of multiple nonsense mutations, some are still active and may play a role in human diseases, especially the HERV type K (HERV-K) family.3-5 Transactivation of HERV-K family members was observed in a variety of human cancers. For instance, the expression of the HERV-K envelope (Env) protein in malignant breast cancer cell lines was substantially higher than nonmalignant breast cells, and anti-HERV-K-specific monoclonal antibodies inhibited growth, and induced apoptosis of breast cancer cells in vitro and in vivo.6 Targeting HERV-K Env by CRISPR/Cas9 significantly downregulated prostate cancer cell line expression of proto-oncogene SF2/ASF as well as the RAS pathway.7 The downregulation of HERV-K expression in pancreatic adenocarcinoma cells impaired tumor functions through RAS–ERK–RSK pathways in several cell lines and the tumor formation was reduced in mouse xenograft models treated with HERV-K Env targeting shRNA.8 Interestingly, Np9, a 9-kDa protein translated from the HERV-Kenv reading frame, has found as an oncogenic protein and present in a variety of tumors and transformed cells.9, 10 Furthermore, Np9 has been reported to not only activate the ERK, Akt and Notch1 pathways but also to upregulate β-catenin, which are essential for survival of some cancer stem cells.9
Kaposi's Sarcoma-associated herpesvirus (KSHV) represents a principal causative agent of several cancers arising in patients with compromised immune systems, such as Kaposi's Sarcoma (KS) and primary effusion lymphoma (PEL).11 Despite the reduced incidence of KS in the era of combined antiretroviral therapy (cART) for HIV infection, KS still remains the most common acquired immunodeficiency syndrome (AIDS)-associated tumor and a leading cause of morbidity and mortality in this setting.12 Our recent data indicated that KSHVde novo infection of primary endothelial cells and/or KSHV + PEL tumor cells displayed much higher levels of HERV-K env transcripts when compared to UV-inactivated KSHV infected cells or virus-negative lymphoma cells, respectively.13 Further functional assays indicated that activation of Np9 expression was responsible for enhancing cell invasiveness and anchorage-independent growth of KSHV+ cells. We also demonstrated the upregulated expression of Np9 within KS tumor tissues when compared to adjacent normal tissues from AIDS-KS patients without any treatment.13 In the current study, we for the first time reported that ectopic expression of Np9 protein was able to induce DNA damage response (DDR) from host cells especially through upregulation of γH2AX and other DDR-related proteins. Interestingly, we found that direct knockdown of Np9 by RNAi in KSHV-infected cells effectively reduced LANA expression, one of KSHV-encoded major latent oncoproteins in vitro and in vivo.
2 MATERIALS AND METHODS
2.1 Cell culture and reagents
Human embryonic kidney (HEK) 293A cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. KSHV long-term-infected telomerase-immortalized human umbilical vein endothelial cells (TIVE-LTC) were kindly provided by Dr. Rolf Renne (University of Florida) and cultured as previously described.14 All chemicals were purchased from Sigma-Aldrich.
2.2 Cell proliferation and apoptosis assays
Cell proliferation was assessed using the WST-1 Assay (Roche). After treatment, 10 μL/well of WST-1 reagent was added to 96-well plates and incubated for 3 h at 37°C in 5% CO2. Absorbance at 490 nm was measured using a microplate reader. Flow cytometry with the FITC-Annexin V/propidium iodide (PI) Apoptosis Detection Kit I (BD Pharmingen) was employed for quantitative apoptosis assessment, analyzed on a FACS Calibur 4-color flow cytometer (BD Bioscience).
2.3 Immunoblotting
Total cell lysates (30 µg) were resolved by 10% SDS–PAGE, transferred to nitrocellulose membranes, and incubated with 100–200 µg/mL of antibodies provided in DNA Damage Antibody Sampler Kit (Cell Signaling, #9947). GAPDH served as the loading control (Cell Signaling). LANA antibody was purchased from Abcam. HERV-K Np9 antibody was kindly provided by Dr. Friedrich A. Grasser from Universitatsklinikum des Saarlandes, Germany.10 Immunoreactive bands were identified using an enhanced chemiluminescence reaction (Perkin-Elmer) and visualized by autoradiography.
2.4 CometAssay
The DNA damage was evaluated by using the reagent kit for single cell gel electrophoresis assay/CometAssay (Trevigen), according to the manufacturer's instructions. The slides were viewed by using epifluorescence microscopy.
2.5 Plasmid transfection and RNA interference
Cells were transfected with control vectors, pDsred-NP9, pSG5-NP9 (both are kindly provided by Dr. Friedrich A. Grasser),10 in 12-well plates using Lipofectamine 3000 (Invitrogen). To establish stable HERV-K knockdown cells, we used Dharmacon SMARTvector LentiviralNp9-shRNA and a nonsilencing (n)-shRNA as a negative control.
2.6 Immunofluorescence assays
Briefly, HEK293A cells were seeded in two-well chamber slides (Nunc) and transfected with the pDsred-NP9 vector for 48 h. After that, slides were incubated in 1:1 methanol–acetone at 20°C for fixation and permeabilization and then with a blocking reagent (10% normal goat serum, 3% bovine serum albumin, and 1% glycine) for an additional 30 min. Cells were then incubated for 1 h at 25°C with an anti-γH2AX antibody (Cell Signaling) followed by a secondary antibody conjugated to Alexa 488 (Invitrogen). For identification of nuclei, cells were subsequently counterstained with 0.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; Sigma) in 180 mM Tris-HCl (pH 7.5). Slides were washed once in 180 mM Tris-HCl for 15 min and prepared for visualization using a Leica TCPS SP2 AOBS microscope.
2.7 KS-like nude mouse model
5 × 105 TIVE-LTC cells in 50 µL PBS plus 50 µL growth factor-depleted Matrigel (BD Biosciences) were together injected subcutaneously into the flanks of nude mice, 6-8-week-old, male (Jackson Laboratory). At the end of experiment, the tumors were excised for immunohistochemistry analyses of LANA expression as described previously.15 Tissue slides were then scanned with an Aperio CS2 digital pathology scanner. Images were obtained with Aperio ImageScope software (Leica). The percentage of DAB stained pixels were determined by analyzing the raw images with the QuPath software (version 0.2.3).16 All protocols were approved by the UAMS Institutional Animal Care and Use Committee (IACUC) in accordance with the national guidelines.
2.8 Statistical analysis
Significance for differences between experimental and control groups was determined using the two-tailed Student's t test (Excel 2016).
3 RESULTS AND DISCUSSION
3.1 Ectopic expression of HERV-K Np9 reduces the growth of host cells
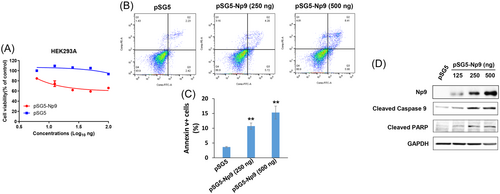
We found that ectopic expression of Np9 protein by transiently transfection of recombinant plasmids caused the inhibition of growth of HEK293A cells (Figure 1A) as well as other cells such as HeLa (data not shown) in a dose-dependent manner. Flow cytometry analysis indicated that overexpression of Np9 induced host cell apoptosis (Figure 1B,C). Western blot results confirmed that the expression of several apoptosis markers such as cleaved Caspase 9 and PARP was upregulated by Np9 (Figure 1D).
3.2 HERV-K Np9 induces and/or enhances DNA damage in host cells
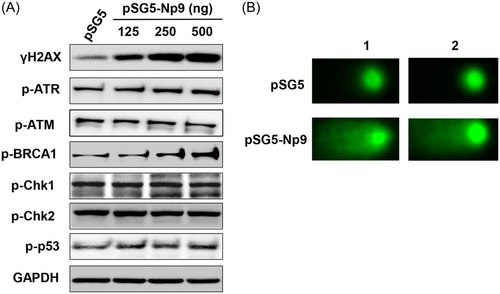
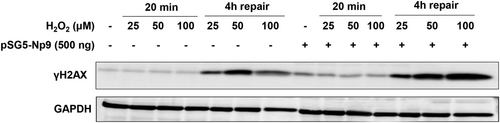
Interestingly, we found that ectopic expression of Np9 protein also caused obvious DDR in host cells. By using DNA Damage Antibody Sampler Kit from Cell Signaling, we detected and screened the expression of several major DDR-related proteins. We found that Np9 increased γH2AX (one of DNA damage markers) together with phosphor-ATR, phosphor-BRCA1, but not affecting phosphor-ATM, phosphor-Chk1/Chk2 or phosphor-p53 (Figure 2A). The CometAssay results confirmed that overexpression of Np9 induced obvious DNA damage (comet tails) from HEK293A cells (Figure 2B). We next treated HEK293A cells with a DNA damage inducer, H2O2, for 20 min,17 then incubated cells for 4 h allowing them for repair. As shown in Figure 3, we found that overexpression of Np9 obviously interfered with DNA repair process in host cells with higher levels of γH2AX than the controls. Together, our data for the first time show that overexpression of HERV-K Np9 protein can induce DDR from host cells, which may cause or accumulate genomic instability, enhancing the tumorigenesis even partially reducing cell growth.
3.3 Directly targeting Np9 reduces KSHV LANA expression in vitro and in vivo
KSHV de novo infection has been found to upregulate the level of γH2AX, which interacts with LANA and contributes to LANA-mediated episome maintenance.18, 19 Furthermore, H2AX knockdown reduces the expression of LANA and viral genome copies, suggesting that γH2AX has a role in latent gene expression and establishment of KSHV latency.19 As we already showed the interaction between LANA and Np9 in the nuclear,13 here we found that most of Np9 proteins colocalized with γH2AX in the nuclear of Np9-transfected cells by using immunofluorescence assays (Figure 4A). Interestingly, we found that stably knockdown of Np9 by shRNA from TIVE-LTC (a KSHV long-term-infected immortalized endothelial cell line) 14 significantly reduced the expression of LANA as well as c-Myc (Figure 4B), the latter has been reported to directly interact with LANA.20 As a proto-oncogene, c-Myc has been found to induce DNA damage, increase reactive oxygen species (ROS), and mitigate p53 function.21 In addition, the in vivo study also reveals an essential role for endogenous c-Myc in signaling DNA damage-induced apoptosis through the control of the p53 function.22
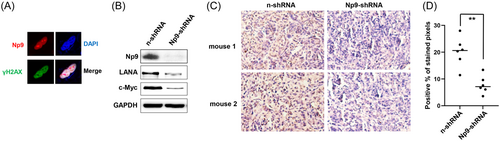
By using a KS-like xenograft model, we recently reported that nude mice subcutaneously injected with Np9-shRNA stably knockdown TIVE-LTC formed much smaller tumors, when compared to mice injected with control shRNA (“n-shRNA”) cells at 30 days pi.13 Here we re-examined the slides and found that stably knockdown of Np9 greatly reduced LANA expression in tumor tissues (Figure 4C). Our quantification analysis confirmed ~60% of reduction of LANA expression in tumor tissues from Np9 stably knockdown cells injected mice when compared to those from the control group (Figure 4D).
In summary, our findings discover the association between HERV-K Np9 protein and DDR in host cells, representing a novel mechanism for Np9 oncogenic functions. Further, we found that targeting Np9 effectively reduced LANA expression, one of KSHV-encoded major latent oncoproteins in vitro and in vivo, which may represent a novel direction to develop therapeutic strategy.
AUTHOR CONTRIBUTIONS
Conception and design: Lu Dai, Zhiqiang Qin. Development of methodology: Zhen Lin. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Jungang Chen, Jiaojiao Fan, Zhen Lin, Lu Dai. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Lu Dai, Zhiqiang Qin. Writing, review, and/or revision of the manuscript: Lu Dai, Zhiqiang Qin. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Zhen Lin. Study supervision: Zhiqiang Qin.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health R03DE031978, and the Arkansas Bioscience Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000. Z. L. was supported by a National Cancer Institute grant R01CA261258, a National Institute of General Medical Sciences COBRE grant P20GM121288, a U.S.-Japan Cooperative Medical Sciences Program Collaborative Award from the National Institute of Allergy and Infectious Diseases and CRDF Global (grant number DAA3-19-65602-1), a Ladies Leukemia League research grant, a Tulane school of medicine faculty research pilot grant, and a Carol Lavin Bernick faculty grant. Funding sources had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




