DENV-1 genotype V circulation during the nonepidemic period in the Northeast of São Paulo State endemic area
Dengue virus (DENV), one of the most critical arboviruses, has caused extensive outbreaks in Latin America, particularly in Brazil. One of the lessons learned during the Covid-19 pandemic was that it is possible to establish real-time genomic surveillance to acquire information regarding the origin and spread of different viral variants. However, molecular surveillance of DENV genotypes during outbreaks or the prediction of their emergence remains a considerable challenge in the DENV epidemiology in Brazil.
All four DENV serotypes (DENV-1–4) have been circulating in Brazil over the last 10 years. A systematic literature review showed that the predominant serotypes vary from region to region over a period.1 Between 2000 and 2009, DENV-1–3 circulated simultaneously with the entry of DENV-4 in 2010. Since then, all serotypes have co-circulated. DENV-4 was the most prevalent serotype in 2012, particularly in the northern, northeastern, and southeastern regions. DENV-1 was dominant from 2014 to 2016, whereas DENV-2 was prevalent in 2018 in the entire country.1
The Ribeirão Preto region, located in the interior of São Paulo State, is hyperendemic for DENV. The region has experienced multiple and fluctuating trends in DENV outbreaks,2 characterized by an increase in cases in 2013, 2015, 2016, 2019, 2020, and 2022, as illustrated in Figure 1A. Of particular concern is the severe outbreak in 2016, in which the co-circulation of dengue and Zika viruses was observed, and over 35 000 cases were reported.3, 4 The continuous presence of DENV in a population can serve as a model for investigating viral evolution over time as the virus continues to replicate and evolve, leading to the emergence of novel viral variants. Therefore, monitoring DENV circulation during nonepidemic periods is crucial for predicting novel epidemics and the potential emergence of emerging viral variants.
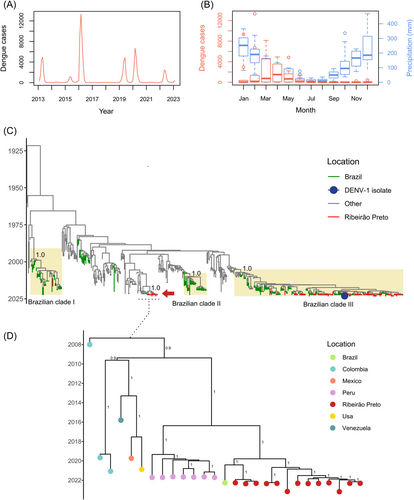
Patients were examined by medical professionals at a public health unit for the presence of dengue symptoms. Plasma samples were collected from 20 patients suspected of DENV infection. Patients referred for blood collection and NS1 antigen detection were invited to participate in the study and signed a consent form. Active surveillance ranged from October 2022 to January 2023, comprising the nonepidemic period when the incidence of confirmed dengue cases was low and coincided with the rainy season (Figure 1B). During this period, Ribeirão Preto reported 210 confirmed DENV cases, corresponding to an incidence of 30 per 100 000 inhabitants. Among the tested participants, 25% were female, with a mean age of 39 years (range: 20–60 years). All participants presented one of the following symptoms: fever, headache, myalgia, arthralgia, retroorbital pain, or rash. Additionally, 65% of the patients reported previous dengue infections.
Two tested samples were DENV-1 PCR-positive (GeneFinder™ DENV Typing RealAmp Kit, Osang Healthcare), but only one was NS1 antigen-positive (ELISA Ag, Bio-Rad). Comprehensive details of the demographics, laboratory findings, and clinical data of the study participants are summarized in Supporting Information S1: Table 1. Of the 20 DENV PCR tests conducted in this study, a positivity rate of 10% (n = 2) was observed, corresponding to a statistical power of 74%, which aligned with the officially reported proportional positivity between suspected and confirmed cases. Based on the cycle threshold value (Ct = 20.7), only one sample was submitted to whole genome sequencing using adapted Illumina COVIDSeq protocol (Illumina). DENV-specific primers were used for the amplification step (Supporting Information S1: Table 2). Amplification and library preparation steps were performed according to the manufacturer's instructions. Phylogenetic analysis conducted during the epidemic period of 2022 (March to May) revealed that there were two DENV1 clades circulating in Ribeirão Preto. The DENV-1 sequence obtained in this study clustered with Brazilian samples within clade III genotype V (Figure 1C, blue dot), which is the predominant clade currently circulating in Ribeirão Preto. Additionally, 13 samples of DENV-1 (red arrow) from the same location clustered in a separate DENV-1 clade, together with strains from South America (Figure 1D). Notably, there was no evidence of a recent common ancestor among the identified clades.
Close monitoring of cryptically circulating DENV genotypes in the study area may reveal patterns that can help predict future epidemics, especially when considering genotypes that have not been circulating in this region. Over the past 2 years, both DENV-1 and DENV-2 have been responsible for epidemics in the Ribeirão Preto region. In 2019, an outbreak of DENV-2 Asian/American genotype was registered, which possibly originated from the Caribbean.4 DENV-3 circulated from 2003 to 20086 and from 2010 to 2011,7 except in 2021, when only DENV-1 and DENV-2 were detected.5, 8
In 2022, a cosmopolitan genotype (DENV-2 genotype II) was detected for the first time in Midwestern Brazil.9 Although some Brazilian states have reported the DENV-2 cosmopolitan genotype, its incidence remains lower than DENV-1. There is still no circulation of this genotype in the Ribeirão Preto region; however, cases have been reported in other locations in São Paulo state and have shown a steady spread throughout the country.9
This study highlights the significance of genomic surveillance for DENV. Despite the considerable financial burden, this underscores the urgent need to implement genomic surveillance within affected regions, particularly in developing nations with inadequate healthcare infrastructure. Technical challenges associated with DENV genomic surveillance must also be overcome, given the fragile nature of biological specimens and the duration of viremia. Strategies to improve molecular epidemiological monitoring must be developed to facilitate the detection of emerging DENV genotypes, thereby preventing future epidemics and promptly implementing proactive measures to control and prevent DENV transmission. Studying the evolution could provide insights into disease severity, vaccine development, and transmissibility.
AUTHOR CONTRIBUTIONS
Simone Kashima: Conceptualization; data curation; formal analysis; writing original and draft preparation; funding acquisition. Antonio Jorge Martins: Conceptualization. Denise Bergamaschi Giomo: Conceptualization. Luzia Márcia Romanholi Passos: Conceptualization; Writing—review and editing. Paula Marilia Afonso Torres: Conceptualization. Danielle Cristina Dacanal Gentil: Conceptualization. Erika Aparecida Catoia: Conceptualization. Natalia do Carmo Chiquito: Conceptualization; methodology. Alessandra Paula Silva Soares Medeiros: Conceptualization. Elaine Vieira Santos: Methodology. Debora Glenda Lima de La-Roque: Methodology; data curation; formal analysis; writing original and draft preparation. Renata Aparecida Machado Oliveira: Methodology. Evandra S. Rodrigues: Methodology. Marta Giovanetti: Writing—review and editing. Vagner Fonseca: Data curation; formal analysis. Svetoslav Nanev Slavov: Writing—review and editing. Aparecida Y. Yamamoto: Writing—review and editing. Rodrigo Tocantins Calado: Writing—review and editing. Maria Carolina Elias: Writing—review and editing; funding acquisition. Sandra Coccuzzo Sampaio: Writing—review and editing; funding acquisition. Luiz Carlos Junior Alcantara: Writing—review and editing; funding acquisition. Dimas T. Covas: Writing—review and editing; funding acquisition.
ACKNOWLEDGMENTS
This research was funded by the Fundação de Amparo à Pesquisa do Estado de São Paulo FAPESP (grants 2013/08135-2; 2021/11944-6; 2022/16349-1); Conselho Nacional de Pesquisa e Desenvolvimento, CNPq (465539/2014-9), FUNDHERP. We would also like to thank all authors who kindly deposited and shared the genomes in GenBank (NCBI). MG was funded by PON “Ricerca e Innovazione” 2014–2020. This work was partially supported by the National Institutes of Health, USA, grant U01 AI151698 to the United World Arbovirus Research Network (UWARN). The authors would like to acknowledge the Global Consortium to Identify and Control Epidemics—CLIMADE (T. O., L. C. J. A., E. C. H., J. L., M. G.) (https://climade.health/).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The study was approved by the Ethics Committee of the Hospital das Clinicas de Ribeirao Preto, Faculty of Medicine, under Process CAAE:59073722.0.0000.5440. Written informed consent was obtained from all the participants.
Open Research
DATA AVAILABILITY STATEMENT
The genomes analyzed in the present study were obtained from the GenBank database and are available under the accession number: OQ872854.




