Evolution of SARS-CoV-2 variants of concern over a period of Delta and Omicron cocirculation, among patients hospitalized for COVID-19 in an Italian reference hospital: Impact on clinical outcomes
Annalisa Mondi, Ilaria Mastrorosa, Enrico Girardi, and Andrea Antinori contributed equally to this study.
Abstract
Despite the higher transmissibility of Omicron Variant of Concern (VOC), several reports have suggested lower risk for hospitalization and severe outcomes compared to previous variants of SARS-CoV-2. This study, enrolling all COVID-19 adults admitted to a reference hospital who underwent both the S-gene-target-failure test and VOC identification by Sanger sequencing, aimed to describe the evolving prevalence of Delta and Omicron variants and to compare the main in-hospital outcomes of severity, during a trimester (December 2021 to March 2022) of VOCs' cocirculation. Factors associated with clinical progression to noninvasive ventilation (NIV)/mechanical ventilation (MV)/death within 10 days and to MV/admission to intensive care unit (ICU)/death within 28 days, were investigated through multivariable logistic regressions. Overall, VOCs were: Delta n = 130/428, Omicron n = 298/428 (sublineages BA.1 n = 275 and BA.2 n = 23). Until mid-February, Delta predominance shifted to BA.1, which was gradually displaced by BA.2 until mid-March. Participants with Omicron VOC were more likely to be older, fully vaccinated, with multiple comorbidities and to have a shorter time from symptoms' onset, and less likely to have systemic symptoms and respiratory complications. Although the need of NIV within 10 days and MV within 28 days from hospitalization and the admission to ICU were less frequent for patients with Omicron compared to those with Delta infections, mortality was similar between the two VOCs. In the adjusted analysis, multiple comorbidities and a longer time from symptoms' onset predicted 10-day clinical progression, while complete vaccination halved the risk. Multimorbidity was the only risk factor associated with 28-day clinical progression. In our population, in the first trimester of 2022, Omicron rapidly displaced Delta in COVID-19 hospitalized adults. Clinical profile and presentation differed between the two VOCs and, although Omicron infections showed a less severe clinical picture, no substantial differences for clinical progression were found. This finding suggests that any hospitalization, especially in more vulnerable individuals, may be at risk for severe progression, which is more related to the underlying frailty of patients than to the intrinsic severity of the viral variant.
1 INTRODUCTION
Since the outbreak of coronavirus disease 2019 (COVID-19) in late 2019, the world has witnessed successive waves of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections driven by the emergence of new variants of concern (VOCs), characterized by increased transmissibility, which have posed major clinical and epidemiological challenges.1, 2 So far, five SARS-CoV-2 variants have been identified as VOCs by the World Health Organization (WHO): Alpha (B.1.1.7), the first VOC to be identified in late 2020; Beta (B.1.351); Gamma (P.1); Delta (B.1.617.2), designated as a VOC in the spring of 2021 and characterized by greater contagiousness and severity compared to previous strains; and Omicron (B.1.1.529, including all sublineages and descendant lineages).3-9
The Omicron variant was first identified in November 2021 in South Africa and subsequently spread worldwide rapidly displacing the Delta variant and triggering for a new surge of SARS-CoV-2 infections which is still ongoing.10, 11 The Omicron sublineage BA.1 drove the first omicron-related wave from mid-to-late December 2021 to the end of January 2022 and was subsequently replaced by the Omicron sublineage BA.2, responsible for the second omicron wave, which became dominant from mid-February 2022.11 The Omicron VOC showed higher transmissibility and an increased risk of escape from infection- or vaccination-induced immunity compared to previous variants.12-14 This growth advantage seemed to be explained by the presence of a significative number of mutations involving the spike gene of the virus, some of whom are known to lead to higher viral binding affinity and lower response to previous immunity.15-18 Nevertheless, in addition to these aspects of concerns, several studies have reported a reduced risk for hospitalization and severe outcomes of Omicron compared to Delta infections.19-24 However, whether this lower degree of severity is related to an inherent characteristic of the virus or an increase in immunity in the population over time remains to be clarified. Despite the lower severity of Omicron infections reported in general population, comparisons of the clinical profile and the risk for clinical progression between the two VOCs in subjects hospitalized for SARS-CoV-2 infections are still limited.25-29 Additionally, the role of no-viral factors as predictors for severe in-hospital outcomes, in the period of Delta-to-Omicron transition, should be better defined, particularly in high vaccine coverage settings, where a high proportion of admissions are associated with, but not caused by, SARS-CoV-2 infection.
We hereby aimed to describe the evolution in prevalence of the Delta and Omicron variants among patients hospitalized for COVID-19 during a 3-month period when both VOCs were co-circulating. We also aimed to compare baseline characteristics and main in-hospital outcomes according to the identified VOC and to assess the predictive factors of clinical progression.
2 MATERIALS AND METHODS
2.1 Study procedures and population
We conducted an observational, prospective, monocenter study including all consecutive patients admitted to the National Institute for Infectious Diseases “Lazzaro Spallanzani,” in Rome, Italy, in both general wards and intensive care unit (ICU), from December 24, 2021 to March 15, 2022, with confirmed COVID-19 diagnosis and the SARS-CoV-2 variant identified.
We considered as baseline the visit performed at the time of hospital admission, which included medical examination, laboratory testing, vital signs recording and selfreported symptoms' assessment. The presence of pneumonia was evaluated by computed tomography (CT) scan of the chest, performed when admitted or immediately before, in the Emergency and Accident Department. Demographics, medical history and data on comorbidities were also collected. Data on vaccination were extracted from the regional register (Anagrafe Vaccinale Regione Lazio) and, if not available, we collected selfreported vaccination status from hospital charts. Participants were defined: (1) unvaccinated, if they did not receive any vaccine dose, or they received only the first dose of a two-dose series less than 14 days earlier; (2) partially vaccinated, if they received only the first dose of a 2-dose series more than 14 days earlier, or if they completed the vaccine schedule less than 14 days earlier; (3) recent fully vaccinated or boosted, if they completed the vaccine schedule between 14 and 120 days earlier, or if they received the booster dose more than 7 days before the hospital admission; (4) waned fully vaccinated or unboosted, if they completed the vaccine schedule more than 120 days earlier, without having received the booster dose or if they had the booster dose less than 7 days before the hospital admission.
Participants were followed up to Day 10 to assess any prescribed medications (steroids, remdesivir, monoclonal antibodies, and immunotherapy), the use of noninvasive ventilation (NIV), ICU admission with mechanical ventilation (MV), and in-hospital death; clinical progression with ICU admission, MV and in-hospital death were also evaluated up to day 28.
All the data were collected by a trained staff and anonymously recorded into an electronic database.
2.2 Virological assessment
Diagnosis of SARS-CoV-2 infection was confirmed by the detection of viral RNA in nasopharyngeal swab (NPS), or bronchoalveolar lavage (BAL) by Real-Time polymerase chain reaction (RT-PCR) targeting both the viral RdRp- and N-genes resulting in a single cycle threshold (Ct) value indicative of the detection of SARS-CoV-2 RNA (RealTime SARS-CoV-2 Assay; Abbott) on Alinity platform (Abbott).
The “TaqPath™ COVID-19 CE-IVD RT-PCR Kit” (ThermoFisher Scientific), a multiplexed assay that contains three primer/probe sets specific to ORF1ab, N gene and S gene of SARS-CoV-2, was used as S Gene Target Failure (SGTF) test on extracted RNA from NPS or BAL of hospitalized COVID-19 patients. Nucleic acid extraction was performed by QiaSymphony automatic extractor using a DSP Virus/Pathogen Kit (both from QIAGEN).
By kit instruction, the cycle threshold cut-off value for clinical target was ≤37. Since the Omicron BA.1 variant, in contrast to the Delta and Omicron BA.2, is characterized by a deletion at 69 and 70 amino acid positions of the spike protein, this test was able to detect the presence or absence of the 69-70 deletion. The failure of the probe targeting the S gene, in presence of ORF1ab and N gene amplifications, is also referred to as S drop-out. This presence of S drop-out in an otherwise positive PCR test has proven to be a surrogate marker for the Omicron variant, sublineage BA.1.13 Subsequently identification of SARS-CoV-2 variants was conducted by Sanger sequencing of the Spike coding gene, as previously reported.30
2.3 Data analysis
The evolution of VOCs' prevalence throughout the study period was weekly calculated, considering Delta, Omicron BA.1, and Omicron BA.2 variants.
Descriptive characteristics at hospital admission were provided using median values and interquartile ranges (IQR) for continuous variables, and frequencies and percentages for categorical variables, and were compared between Delta and Omicron groups using Chi-square test (Fisher's exact test when applicable) for categorical variables and Mann-Whitney test for continuous variables. Similarly, comparison of in-hospital treatments and main clinical outcomes was assessed according to VOC.
Predictive factors of clinical progression within 10 days (short-term clinical progression) and 28 days from hospital admission (long-term clinical progression) were assessed using multivariable logistic regression calculating odds ratios (MLR-OR) and their 95% confidence intervals (95% CI). The following factors were a priori included as covariates in the MLR model: gender, age, number of comorbidities, vaccination status, days from symptoms onset and VOC. Short-term clinical progression was defined as a combinate outcome of NIV, MV, or death occurred within 10 days from hospitalization. Long-term clinical progression was defined as a combinate outcome of ICU admission, MV, or death occurred within 28 days from hospitalization.
In addition, SARS-CoV-2 Ct values on NPS collected within 3 days from hospital admission in patients infected with Delta and Omicron variants were compared between them using the Mann–Whitney test, after stratifying samples according to days elapsed from symptoms onset to NPS (≤7 days vs. >7 days).
Finally, the sequences obtained by Sanger method were aligned using MAFFT software v7.271,31 with Wuhan-Hu-1 reference sequence (accession number: NC_045512.2) and high-quality, full-length genome representatives for main lineages.32, 33 Quality controls on mutations, deletions, and missing nucleotides were performed with BioEdit and MEGAX. Maximum likelihood (ML) phylogenetic analysis was performed with IQ-TREE v.1.6.1234; best tree model was selected using ModelFinder35; the best tree was found performing 5000 bootstrap ultrafast replicates.
Due to the low number of patients with Omicron BA.2 VOC in our population, Omicron sublineages BA.1 and BA.2 were merged in a single group named Omicron in all the statistical analysis. The descriptive analysis of patients' baseline characteristics was performed also by differentiating the three VOCs (Delta, Omicron BA.1, and Omicron BA.2) using Chi-square test (Fisher's exact test when applicable) for categorical variables and Kruskall–Wallis test for continuous variables and was added as Supporting Information.
A p-value less than 0.05 indicated conventional statistical significance. All statistical analysis was performed using IBM SPSS Statistics version 28 (IBM Corp.) and STATA release 17 (StataCorp. 2021. StataCorp LLC).
3 RESULTS
Over the study period, 532 patients with a diagnosis of COVID-19 were admitted at our Institute. Among them, 104 were excluded from the analysis because of failure to identify the viral variant by sequencing (n = 97) or because of lack of complete patients' data (n = 7). A total of 428 subjects were included in the study: 130 infected with Delta VOC and 298 with Omicron VOC (n = 275 sublineage BA.1 and n = 23 sublineage BA.2) (Figure 1).
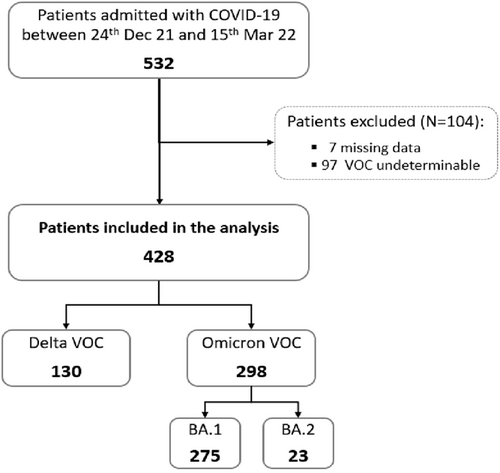
Study flow chart. Dec, December; Mar, March; VOC, variant of concern; N, number.
Concerning the variant screening by SGTF test, after Sanger sequencing identification, as expected, most of the subjects infected with Omicron BA.1 variant tested positive (263/275, 95.6%), while most of the patients infected with Delta variant (122/130, 93.8%) and all those with Omicron BA.2 were negative. For some samples, the TaqPath test gave inaccurate or inconclusive results (negative for 7/275 [2.5%] Omicron BA.1; positive for 4/130 [3.1%] and indeterminate for 1/130 [0.8%] Delta), probably due to high Ct values. Finally, in the case of few participants harboring Delta (3/130, 2.3%) and Omicron BA.1 (5/275, 1.8%), the SGTF test was missed.
3.1 Evolution of variants of concern prevalence
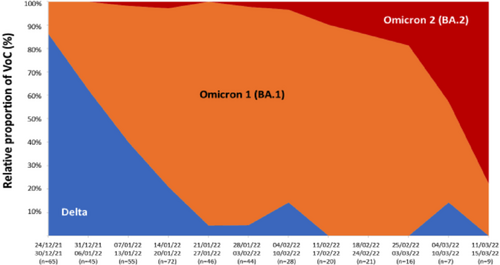
At the beginning of the study period, Delta was the dominant circulating variant in our population, accounting for about 85% of SARS-CoV-2 infections, while Omicron BA.1 was responsible for the remaining 15% of cases. In the following weeks, the proportion of cases due to the Omicron variant increased sharply to constitute more than 90% of the circulating VOCs at the end of January 2022. Omicron BA.1 was the predominant sublineage (>90%) until mid-February 2022, then it was gradually displaced by Omicron BA.2, which emerged in our population in the first week of January 2022 and reached 70% of the circulating variants by the end of the study period (Figure 2).
3.2 Patients' characteristics at hospital admission
The main characteristics at hospital admission are shown in Table 1. Briefly, the median age of the study population was 67 years (IQR 55-79) and 40.7% were women. The majority (79%) of patients had at least one underlying concomitant disease. At hospital admission, which occurred within a median time of 5 days (IQR: 3–9) from symptoms' onset, 85.5% of subjects had evidence of lung involvement at the CT scan.
| Characteristics | VOC | Total (n = 428) | p-valuea | |
|---|---|---|---|---|
| Delta (n = 130) | Omicron (n = 298) | |||
| Females, n (%) | 64 (49.2) | 110 (36.9) | 174 (40.7) | 0.017 |
| Age at hospital admission (years), median (IQR) | 62 (51–72) | 69 (56–80) | 67 (55–79) | <0.001 |
| Days from symptoms' onset, median (IQR) | 7 (5–10) | 5 (2–8) | 5 (3–9) | <0.001 |
| Comorbidities, n (%) | ||||
| Diabetes | 21 (16.2) | 52 (17.5) | 73 (17.1) | 0.743 |
| Hypertension | 55 (42.3) | 136 (45.6) | 191 (44.6) | 0.524 |
| Obesity (BMI > 30) | 26 (20.0) | 54 (18.1) | 80 (18.7) | 0.647 |
| Cardiovascular disease | 22 (16.9) | 79 (26.5) | 101 (23.6) | 0.032 |
| Chronic respiratory disease | 10 (7.7) | 61 (20.5) | 71 (16.6) | 0.001 |
| Renal impairment | 3 (2.3) | 18 (6.0) | 21 (4.9) | 0.100 |
| Immunosuppression | 3 (2.3) | 25 (8.4) | 28 (6.5) | 0.019 |
| Cancer | 14 (10.8) | 53 (17.8) | 67 (15.7) | 0.066 |
| Number of comorbidities, n (%) | 0.007 | |||
| 0 | 38 (29.2) | 52 (17.5) | 90 (21.0) | |
| 1 | 43 (33.1) | 83 (27.9) | 126 (29.4) | |
| 2 | 20 (15.4) | 71 (23.8) | 91 (21.3) | |
| ≥3 | 29 (22.3) | 92 (30.9) | 121 (28.3) | |
| Vaccine status, n (%) | <0.001 | |||
| Not vaccinated | 92 (70.8) | 122 (40.9) | 214 (50.0) | |
| Partially vaccinated | 4 (3.1) | 14 (4.7) | 18 (4.2) | |
| Recent fully vaccinated/boosted | 10 (7.7) | 89 (29.9) | 99 (23.1) | |
| Waned fully vaccinated/unboosted | 24 (18.5) | 73 (24.5) | 97 (22.7) | |
| Symptoms, n (%) | ||||
| Fever | 99 (76.7) | 199 (67.2) | 298 (70.1) | 0.049 |
| Cough | 102 (79.1) | 177 (60.2) | 279 (66.0) | <0.001 |
| Rhinorrhea | 8 (6.3) | 30 (10.4) | 38 (9.1) | 0.183 |
| Dyspnea | 86 (78.9) | 149 (56.4) | 235 (63.0) | <0.001 |
| Asthenia | 66 (51.6) | 110 (38.2) | 176 (42.3) | 0.011 |
| Chills | 33 (25.8) | 34 (11.8) | 67 (16.1) | <0.001 |
| PaO2/FiO2, mmHg | ||||
| Median (IQR) | 257 (176–316) | 295 (209–362) | 280 (198–347) | 0.001 |
| ≥300, n (%) | 39 (30.0) | 133 (44.6) | 172 (40.2) | |
| Missing | 2 (1.5) | 27 (9.1) | 29 (6.8) | |
| Laboratory values, median (IQR) | ||||
| Lymphocytes, x103/μL | 0.8 (0.6–1.2) | 0.9 (0.6–1.4) | 0.9 (0.6–1.3) | 0.244 |
| C-reactive protein, mg/dL | 5.3 (2.3–11.1) | 5.2 (1.8–11.3) | 5.2 (1.9–11.2) | 0.672 |
| Neutrophils/Lymphocytes (NLR) | 5.5 (2.5–10.6) | 4.7 (2.5–10.4) | 4.8 (2.6–10.6) | 0.239 |
| Pneumonia, n (%) | ||||
| No | 8 (6.2) | 54 (18.1) | 62 (15.5) | 0.001 |
| Yes | 122 (93.8) | 244 (81.9) | 366 (85.5) | |
- Abbreviations: BMI, body mass index; IQR, interquartile range; n, number of study participants; PaO2/FiO2, ratio of arterial oxygen partial pressure to fractional inspired oxygen; VOC, variant of concern.
- a Chi-square or Mann−Whitney test as appropriate. p < 0.05, indicating conventional statistical significance, are reported in boldface.
Patients infected with the considered VOCs differed for most of the main baseline characteristics. Particularly, patients diagnosed with Omicron VOC compared to those with Delta were more likely to be older (69 years [IQR: 56–80] vs. 62 years [IQR: 51–72], p < 0.001) and to have multiple comorbidities (patients with at least one comorbidity: 82.5% vs. 70.8%, p = 0.007). Hypertension was the most common concomitant disease, regardless the type of VOC. Cardiovascular and chronic respiratory diseases and immunosuppression were significantly more common in patients harboring Omicron (26.5% vs. 16.9%, p = 0.032, 20.5% vs. 7.7%, p = 0.001 and 8.4% vs. 2.3%, p = 0.0019, respectively). Concerning the vaccine status, most patients admitted with Delta VOC infection were not vaccinated (70.8%), whereas more than half of patients with Omicron infection (54.4%) received a full vaccine schedule (p < 0.001). Notably, 29.9% of patients in the Omicron group received the full schedule less than 120 days before the admission or the booster dose.
Participants diagnosed with Delta infection, tended to have a more severe clinical presentation compared to those with Omicron with a higher rate of systemic symptoms (fever 76.7% vs. 67.2%, p = 0.049; chills 25.8% vs. 11.8%, p < 0.001; asthenia 51.6% vs. 38.2%, p = 0.011) and respiratory symptoms (dyspnea 78.9% vs. 56.4%, p < 0.001; cough 79.1% vs. 60.2%, p < 0.001). Additionally, at hospital admission, individuals in the Delta group had a longer time elapsed from symptoms' onset (median days [IQR]: 7 5-10 vs. 5,2-8 p < 0.001), a higher rate of respiratory failure (median PaO2/FiO2 [IQR]: 257 mmHg [176–316] vs. 295 mmHg [213–352], p = 0.001) and lung involvement at the CT scan (93.8% vs. 81.9%, p = 0.001) than patients in the Omicron group.
Looking at baseline characteristics after dividing the Omicron group into Omicron BA.1 and Omicron BA.2 subgroups, we observed that patients in the Omicron BA.2 subgroup were more likely to have multimorbidity (>40% of patients with more than three underlying comorbidities), to have been recently vaccinated/boosted (60.9% of patients). In contrast, they had lower rates of systemic/respiratory symptoms and pulmonary involvement on CT scan (73.9%) and a lower degree of respiratory failure (PaO2/FiO2 376 [IQR: 180–390]) (Supporting Information: Table S1).
3.3 Treatment approaches and in-hospital outcomes
Slightly less than half of our study population (40.6%) received specific treatments for SARS-CoV-2 infection during hospitalization. As expected, the type of antiviral therapy varied according to VOC with a higher number of Remdesivir treatment in the Omicron compared to Delta group (38.3% vs. 22.3%, p = 0.001) (Table 2) and a significantly greater number of treatments with monoclonal antibodies in patients with Delta than those with Omicron infection (13.9% vs. 4.4%, p < 0.001). Of note, 23 subjects (9 with Delta and 14 with Omicron variants) received monoclonal antibodies before hospital admission. A significantly higher proportion of patients infected with Delta infection received steroids (90.8% vs. 71.4%, p < 0.001), whereas no difference in the use of immunotherapy was observed between the groups.
| Characteristics | VOC | Total (N = 428) | p-valuea | |
|---|---|---|---|---|
| Delta (N = 130) | Omicron (N = 298) | |||
| Therapeutic approaches, n (%) | ||||
| Steroids | 118 (90.8) | 212 (71.4) | 330 (77.1) | <0.001 |
| Remdesivir | 29 (22.3) | 114 (38.3) | 143 (33.4) | 0.001 |
| Monoclonal Antibodiesb | 18 (13.9) | 13 (4.4) | 31 (7.2) | <0.001 |
| Immunotherapy | 16 (12.3) | 22 (7.4) | 38 (8.9) | 0.099 |
| Length of hospital (days), median (IQR) | ||||
| 16.5 (10–24) | 16 (9–25) | 16 (9–24) | 0.796 | |
| Clinical outcomes at 10 days from hospital admission, n (%) | ||||
| NIV | 54 (41.5) | 91 (30.5) | 145 (33.9) | 0.027 |
| MV | 22 (16.9) | 33 (11.1) | 55 (12.9) | 0.096 |
| Death | 8 (6.2) | 18 (6.0) | 26 (6.1) | 0.964 |
| ICU | 28 (21.5) | 38 (12.8) | 66 (15.4) | 0.021 |
| NIV/MV/Death | 55 (42.3) | 99 (33.2) | 154 (36.0) | 0.072 |
| Clinical outcomes at 28 days from hospital admission, n (%) | ||||
| MV | 25 (19.2) | 34 (11.4) | 59 (13.8) | 0.031 |
| Death | 15 (11.5) | 34 (11.4) | 49 (11.5) | 0.969 |
| ICU | 28 (21.5) | 40 (13.4) | 68 (15.9) | 0.035 |
| ICU/MV/Death | 29 (22.3) | 56 (18.8) | 85 (19.9) | 0.402 |
- Abbreviations: ICU, intensive care unit; IQR, interquartile range; MV, mechanical ventilation; n, number of study participants; NIV, noninvasive ventilation; VOC, variant of concern.
- a Chi-square or Mann−Whitney test as appropriate. p < 0.05, indicating conventional statistical significance, are reported in boldface.
- b Only the treatment received after hospital admission has been considered.
Concerning the main in-hospital outcomes, the rates of in-hospital death both after 10 and 28 days from hospital admission and the median length of hospital stay did not significantly differ between the groups (Table 2). Conversely, a significant higher proportion of treatments with NIV, within 10 days from hospitalization, was observed among patients diagnosed with Delta (41.5%) compared to those with Omicron (30.5%) (p = 0.027). In addition, higher rates of ICU admission within both 10 and 28 days of hospitalization (21.5% vs. 12.8%, p = 0.021 and 21.5% vs. 13.4%, p = 0.035) and more patients undergoing MV within 28 days (19.2% vs. 11.4%, p = 0.031) were found among patients infected with Delta VOC as compared to those with Omicron (Table 2).
3.4 Predictors of clinical progression
Overall, clinical progression within 10 days from hospital admission, occurred in 154 patients (36%), of whom 55 (42.3%) infected with Delta and 99 (33.2%) infected with Omicron (p = 0.072). At the multivariable logistic regression analysis, short-term clinical progression was predicted by multiple comorbidities with a higher risk as the number of comorbidities increases (MLR-OR 2.29 for two comorbidities, p = 0.024; MLR-OR 3.83 for ≥3 comorbidities, p < 0.001) and a longer time from symptoms onset to hospitalization (MLR-OR 1.96 for >7 vs. ≤7 days, p = 0.004). Conversely, complete vaccination compared to no vaccination lowered the risk of short-term progression (MLR-OR 0.45 for waned fully vaccinated or unboosted vs. not vaccinated, p = 0.07; 0.55 for recent fully vaccinated or boosted vs. not vaccinated, p = 0.039). Being infected with the Omicron compared to Delta VOC showed a trend toward significance as a protective factor for short-term clinical progression (MLR-OR 0.65, p = 0.083) (Table 3).
| Short-term clinical progression | Long-term clinical progression | |||||
|---|---|---|---|---|---|---|
| Cases/Total | MLR-OR (95% CI) | p-value | Cases/Total | MLR-OR (95% CI) | p-valuea | |
| Total | 154/428 | 85/428 | ||||
| Gender | ||||||
| Male | 94/254 | 49/254 | ||||
| Female | 60/174 | 0.77 (0.49–1.20) | 0.253 | 36/174 | 0.95 (0.56–1.60) | 0.844 |
| Age by 10 years of increase | 1.17 (1.00–1.38) | 0.054 | 1.15 (0.95–1.40) | 0.149 | ||
| Number of comorbidities | ||||||
| 0 | 21/90 | 8/90 | ||||
| 1 | 41/126 | 1.75 (0.91–3.35) | 0.093 | 21/126 | 2.08 (0.86–5.03) | 0.104 |
| 2 | 34/91 | 2.29 (1.11–4.72) | 0.024 | 17/91 | 2.41 (0.93–6.24) | 0.070 |
| ≥3 | 58/121 | 3.83 (1.89–7.75) | <0.001 | 39/121 | 4.92 (2.00–12.09) | 0.001 |
| Vaccine status | ||||||
| Not vaccinated | 89/214 | 47/214 | ||||
| Partially vaccinated | 10/18 | 2.18 (0.76–6.25) | 0.146 | 4/18 | 1.09 (0.33–3.65) | 0.890 |
| Recent fully vaccinated/boosted | 29/99 | 0.55 (0.31–0.97) | 0.039 | 16/99 | 0.56 (0.28–1.12) | 0.103 |
| Waned fully vaccinated/unboosted | 26/97 | 0.45 (0.25–0.81) | 0.007 | 18/97 | 0.62 (0.32–1.19) | 0.152 |
| VOC | ||||||
| Delta | 55/130 | 29/130 | ||||
| Omicron | 99/298 | 0.65 (0.40–1.06) | 0.083 | 56/298 | 0.72 (0.41–1.26) | 0.247 |
| Days from symptoms' onset | ||||||
| ≤7 | 87/284 | 56/284 | ||||
| >7 | 67/144 | 1.96 (1.24–3.08) | 0.004 | 29/144 | 1.04 (0.60–1.79) | 0.887 |
- Abbreviations: CI, confidence interval; MLR-OR, multivariable logistic regression-odds ratio; VOC, variant of concern.
- a p < 0.05, indicating conventional statistical significance, are reported in boldface. All variables were included in the model.
Overall, after 28 days from hospital admission clinical progression occurred in 85 (19.9%) patients: 29 (22.3%) in the Delta and 56 (19.3%) in the Omicron group (p = 0.402). After adjusting for the main confounders listed in Methods, having multiple baseline pathologies, particularly more than 2, was the only factor significantly associated with long-term clinical progression (MLR-OR 4.92 compared to no comorbidity, p-value = 0.001). It is worth noting that we did not find any association between the type of VOC and 28-day clinical progression (Table 3).
3.5 Nasopharyngeal viral loads comparison
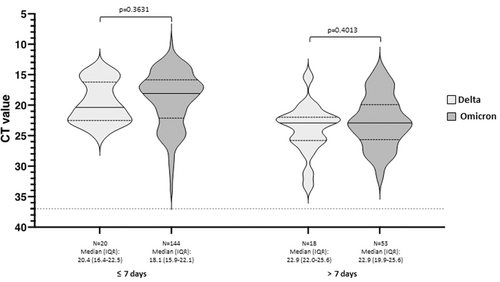
Nasopharyngeal SARS-CoV-2 Ct values obtained within 3 days from hospital admission were available for 235 out of 428 included patients (54.9%, 38 with Delta and 197 with Omicron VOC). Baseline viral loads were compared between patients with Delta and Omicron infection after stratifying according to the time elapsed from symptoms onset. No significant differences in nasopharyngeal Ct values were observed between patients infected with Omicron and Delta VOC at both short (≤7 days) and longer (≥7 days) distance from symptoms onset, as shown in Figure 3.
3.6 Phylogenetic analysis
A specific 523-nucleotide region from SARS-CoV-2 gene S was obtained from respiratory samples of 425 out of 428 patients included in the study. Phylogenetic analysis showed that the obtained sequences clearly segregated in three different genetic groups corresponding to the VOC Delta, Omicron BA.1, and Omicron BA.2, respectively. The sequenced region corresponds to 174 amino acids (aa) of the Spike protein Receptor Binding Domain (from aa position 415 to aa position 588), therefore further analysis was conducted to investigate the occurrence of additional aa mutations within the three genetic groups. As shown in Supporting Information: Figure S1, only six mutations were detected in 425 samples: L425K, I434V, K444I, and F490S were found in seven samples from the Delta group and K417S, L425K, and L455S in four samples from the Omicron BA.1 group. No additional mutations were found in any samples from the Omicron BA.2 group.
4 DISCUSSION
In this prospective observational study, including all patients hospitalized for COVID-19 in the first trimester of 2022 in a COVID-19 Italian reference hospital, we provided a snapshot of the change in clinical features and outcomes of patients admitted during the transition period from Delta to Omicron dominance. In line with national and international epidemiological data,10, 11, 36 in our population, the Omicron VOC sublineage BA.1 rapidly displaced the Delta VOC, becoming the dominant variant by the end of January 2022. Subsequently, it was gradually replaced by the Omicron sublineage BA.2, which became the main circulating variant by the end of the study period.
In our analysis, we observed a different clinical profile of subjects hospitalized with Omicron and Delta variants with the former characterized by older age, greater comorbidity burden, and higher proportion of complete vaccination. In particular, patients infected with Omicron VOC had a median age 7 years higher than those with Delta and more than half of them reported at least two underlying comorbidities at hospital admission as compared to less than 40% of patients infected with Delta. This data is in contrast with previous studies conducted in both outpatient21-23 and inpatient25-27 healthcare settings, which described a younger median age and a similar or lower comorbidity burden of patients with Omicron compared to Delta infection but are in line with recent observations from Belgian28 and British29 cohorts of hospitalized patients. These discrepancies among available studies can be explained by several reasons, such as different approaches and timing of the vaccination campaign, public health measures, and testing strategies. Additionally, the different definitions of COVID-19 hospitalizations among the studies, in which admissions associated with and caused by SARS-CoV-2 infection are not always separated, could contribute to these conflicting results.
Regarding the vaccination status, consistent with previous data from both general population and hospitalized patients,19-29 we found a higher probability of prior full vaccination among Omicron infected individuals, likely due to the mechanism of immune evasion described for this viral variant.12-14 Although this aspect has led to questions about whether the lower severity of Omicron infections may be due to an inherent characteristic of the variant or to the wide vaccination coverage, several studies have confirmed the lower severity of Omicron compared with Delta infections also in the unvaccinated population.22, 24, 37
In our population, the clinical presentation also significantly differed according to the VOC. In fact, participants infected with Omicron were characterized by a reduced rate of systemic and respiratory symptoms and a lower probability of respiratory failure and lung involvement at the CT scan. This finding is consistent with previous studies describing a change in symptoms profile between Omicron and Delta infections with milder manifestations and more upper-airway symptoms in the former compared to the latter.38, 39 The change in viral pathophysiology with the greater tropism for the upper airway of Omicron compared to previous variants could explain this finding. In contrast to recent reports,39 we did not observe a higher symptoms complaint in patients infected with Omicron BA.2 compared to BA.1 sublineage. On the contrary, our results suggest that BA.2 leads to an equally mild course of disease as BA.1, as already described in previous studies.40, 41 However, considering the limited number and the characteristics of patients with Omicron BA.2 in our study, we cannot draw any conclusion
Regarding the main in-hospital outcomes, we did not observe any significant differences in terms of mortality and median length of hospitalization between the two variants. Furthermore, after controlling for potential confounders, Omicron, compared to Delta VOC, was not clearly associated with a different risk of severe disease, both at 10- and 28-day after hospitalization, although a trend toward significance for a lower risk for short-term clinical progression was observed. The discrepancies between our results and data from other studies, which demonstrated a significant lower disease severity for inpatients infected with the Omicron compared to the Delta variant,25-29 could be partially explained by the clinical features of our study population. Interestingly, while several factors such as comorbidities, vaccination status and time from symptoms onset predicted the risk of severe outcomes in the short-term, only a higher number of comorbidities was associated with long-term clinical progression. These data suggest that patients with Omicron compared to Delta VOC, despite an apparently milder clinical presentation, might have similar risk of severe clinical progression which is more related to the infection-associated exacerbations of chronic diseases than to the severity of SARS-COV-2 infection.
Consistently with other reports,22, 42 in the subgroup of patients underwent the nasopharyngeal Ct analysis, we did not observe any statistical difference in viral loads at hospital admission in patients infected with Omicron compared to Delta VOC both for early and for late admissions. Although the limited number of Delta subjects in this subanalysis did not allow any other further stratification (e.g., vaccination status, age), data from previous studies, in which further adjustments were performed, did not show any difference in viral loads between the two VOCs.22, 40 In addition, the phylogenetic analysis conducted on a specific region of the gene S showed a clear segregation of the analyzed sequences into three genetic groups corresponding to the three VOCs (Delta, Omicron BA.1, and Omicron BA.2) with a substantial sequence similarity from isolates belonging to the same group.
Finally, it is worth noting that in our study, the SGTF test correctly detected the majority of participants harboring Omicron BA.1 (96%), Delta (94%), and Omicron BA.2 (100%), allowing us to quickly define the treatment approach. In such a rapidly evolving scenario, the availability of rapid-assays for SARS-CoV-2 variants identification, as the SGTF, may be of crucial utility to promptly adapt appropriate therapeutic strategies, without waiting for the sequencing results. However, these assays identify specific mutations associated with known variants, some of which can be common to different viral strains. The main limit being the inability to recognize recombinant or newly emerged variants, this is way sequencing methods remains the gold standard for the correct characterization of viral genomes.43
Several limitations of our study must be addressed, including its observational nature which is prone to bias due to residual confounding, the lack of data collection on previous infections which does not allow us to assess the potential protective role of prior immunity in the case of breakthrough infections and the failure to evaluate several factors such as immunological response to SARS-CoV-2, inflammatory markers and comedications, which might have affected the disease severity. Although the relatively limited size of our population and the type of subjects analyzed in the study could be considered a bias for a comprehensive comparison between Delta and Omicron infections, they are important for the aim of our analysis, which was primarily to assess differences in clinical presentation at the time of hospital admission and to compare in-hospital prognosis between the two variants among the minority of patients hospitalized because of COVID-19. In addition, the short observation period and the monocentric design of the study might be considered as limitations, as they reduce the generalizability of our findings. However, these factors also represent remarkable strengths which ensure, on the one hand, similar community measures of infection control and levels of vaccination coverage, and, on the other hand, homogeneity in background care, disease management and resource availability. Finally, although the limited number of infections due to Omicron variant BA.2 observed over the study period does not allow us to draw any robust conclusions, at the best of our knowledge, this is one of the few studies which compared infections due to different Omicron sublineages and Delta variant.
In conclusion, our survey showed that, in the first trimester of 2022, Omicron VOC rapidly displaced Delta VOC among patients hospitalized for COVID-19, mirroring the national and international trend of SARS-CoV-2 variant prevalence in the same period. Clinical profile and presentation of individuals hospitalized for SARS-CoV-2 infection differed among the two variants and was milder for those harboring Omicron. Nevertheless, after adjusting for the main confounders, patients infected by Omicron variant, compared to Delta, did not show any substantial lower risk for short- and long-term severe outcomes. The presence of multimorbidity remained the only factor strongly associated with the risk of severe disease suggesting a major role of the exacerbation of the underlying comorbidities in frail subjects, regardless of the viral variant. These findings have several implications, mainly in planning the public health response. In fact, although Omicron VOC is associated to a lower risk of hospital admission,19, 21-23 hospitalized patients enrolled in our study are more likely to be frail and with similar risk of clinical progression and care requirements, as compared to those infected by Delta variant. Therefore, considering the increased contagiousness of more recent VOCs and the preserved effectiveness of vaccination in preventing severe disease,44 efforts for reducing transmission and implementing vaccination, mainly in the more vulnerable population, need to be still guaranteed to avoid a hard-to-sustain pressure on health-care system. Data from different countries with different levels of previous infections and vaccination, are crucial to assess the severity of the omicron variant or new emerging VOCs in a context of continuous viral evolution, to shape public health measures and clinical decisions.
AUTHOR CONTRIBUTIONS
Conceptualization: Annalisa Mondi, Ilaria Mastrorosa, Pierluca Piselli, Claudia Cimaglia, Emanuele Nicastri, Enrico Girardi, and Andrea Antinori. Methodology and formal analysis: Pierluca Piselli and Claudia Cimaglia. Investigation and resources: Annalisa Mondi, Ilaria Mastrorosa, Giulia Matusali, Fabrizio Carletti, Eugenia Milozzi, Elisa Biliotti, Silvia Di Bari, Pierangelo Chinello, Alessia Beccacece, Francesca Faraglia, Pietro Vittozzi, Silvia Mosti, Nardi Tetaj, Giulia Valeria Stazi, Carmela Pinnetti, Marta Camici, Alberto D'Annunzio, Alessandra Marani, Lavinia Fabeni, Cesare Ernesto Maria Gruber, Eliana Specchiarello, and Alberta Villanacci. Data curation: Annalisa Mondi, Ilaria Mastrorosa, Pierluca Piselli, Claudia Cimaglia, Giuseppina Giannico, Eugenia Milozzi, Elisa Biliotti, Silvia Di Bari, Pierangelo Chinello, Alessia Beccacece, Pietro Vittozzi, Silvia Mosti, Nardi Tetaj, and Alberta Villanacci, Sabrina Minicucci. Writing—original draft: Annalisa Mondi and Ilaria Mastrorosa. Writing—review and editing: Pierluca Piselli, Claudia Cimaglia, Giulia Matusali, Fabrizio Carletti, Cesare Ernesto Maria Gruber, Enrico Girardi, and Andrea Antinori. Project administration: Annalisa Mondi, Ilaria Mastrorosa, Emanuele Nicastri, Fabrizio Maggi, Francesco Vaia, Enrico Girardi, and Andrea Antinori. Supervision: Anna Rosa Garbuglia, Stefania Ianniello, Luisa Marchioni, Fabrizio Taglietti, Gianpiero D'Offizi, Fabrizio Palmieri, Emanuele Nicastri, Fabrizio Maggi, Francesco Vaia, Enrico Girardi, and Andrea Antinori. Funding acquisition: Fabrizio Maggi, Francesco Vaia, Enrico Girardi, and Andrea Antinori. All authors contributed to the final version and approved the manuscript for publication.
ACKNOWLEDGMENTS
The authors gratefully acknowledge all the nurse and the clinical and laboratory staff, all the participants, and all the Collaborators Members of the National Institute for Infectious Diseases ReCOVeRI Study Group: Maria Alessandra Abbonizio, Amina Abdeddaim, Elisabetta Agostini, Chiara Agrati, Fabrizio Albarello, Gioia Amadei, Alessandra Amendola, Andrea Antinori, Maria Assunta Antonica, Mario Antonini, Tommaso Ascoli Bartoli, Francesco Baldini, Raffaella Barbaro, Barbara Bartolini, Alessia Beccacece, Rita Bellagamba, Martina Benigni, Nazario Bevilacqua, Gianluigi Biava, Michele Bibas, Elisa Biliotti, Licia Bordi, Veronica Bordoni, Evangelo Boumis, Rosanna Buonomo, Donatella Busso, Marta Camici, Paolo Campioni, Flaminia Canichella, Alessandro Capone, Emanuela Caraffa, Ilaria Caravella, Fabrizio Carletti, Serena Maria Carli, Carlotta Cerva, Roberta Chiappini, Pierangelo Chinello, Maria Assunta Cianfarani, Carmine Ciaralli, Claudia Cimaglia, Nicola Cinicola, Veronica Ciotti, Stefania Cicalini, Francesca Colavita, Angela Corpolongo, Massimo Cristofaro, Salvatore Curiale, Ambrogio Curtolo, Alessandra D'Abramo, Alberto D'Annunzio, Cristina Dantimi, Massimo Cristofaro, Alessia De Angelis, Giada De Angelis, Maria Grazia De Palo, Federico De Zottis, Silvia Di Bari, Virginia Di Bari, Rachele Di Lorenzo, Federica Di Stefano, Gianpiero D'Offizi, Davide Donno, Francesca Evangelista, Lavinia Fabeni, Francesca Faraglia, Anna Farina, Federica Ferraro, Lorena Fiorentini, Andrea Frustaci, Nicoletta Fusco, Marisa Fusto, Vincenzo Galati, Roberta Gagliardini, Paola Gallì, Anna Rosa Garbuglia, Gabriele Garotto, Francesca Gavaruzzi, Ilaria Gaviano, Saba Gebremeskel Tekle, Maria Letizia Giancola, Giuseppina Giannico, Filippo Giansante, Emanuela Giombini, Enrico Girardi, Guido Granata, Maria Cristina Greci, Elisabetta Grilli, Susanna Grisetti, Gina Gualano, Fabio Iacomi, Marta Iaconi, Giuseppina Iannicelli, Stefania Ianniello, Carlo Inversi, Eleonora Lalle, Maria Elena Lamanna, Simone Lanini, Daniele Lapa, Raffaella Libertone, Raffaella Lionetti, Giuseppina Liuzzi, Laura Loiacono, Andrea Lucia, Franco Lufrani, Manuela Macchione, Gaetano Maffongelli, Fabrizio Maggi, Alessandra Marani, Luisa Marchioni, Andrea Mariano, Maria Cristina Marini, Micaela Maritti, Annelisa Mastrobattista, Ilaria Mastrorosa, Giulia Matusali, Valentina Mazzotta, Paola Mencarini, Silvia Meschi, Francesco Messina, Sibiana Micarelli, Eugenia Milozzi, Sabrina Minicucci, Giulia Mogavero, Annalisa Mondi, Marzia Montalbano, Chiara Montaldo, Silvia Mosti, Maria Musso, Michela Nardi, Assunta Navarra, Emanuele Nicastri, Martina Nocioni, Pasquale Noto, Roberto Noto, Alessandra Oliva, Ilaria Onnis, Sandrine Ottou, Claudia Palazzolo, Emanuele Pallini, Fabrizio Palmieri, Giulio Palombi, Carlo Pareo, Virgilio Passeri, Jessica Paulicelli, Federico Pelliccioni, Giovanna Penna, Ada Petrone, Elisa Pianura, Carmela Pinnetti, Maria Pisciotta, Pierluca Piselli, Silvia Pittalis, Maria Maddalena Plazzi, Agostina Pontarelli, Costanza Proietti, Vincenzo Puro, Paolo Migliorisi Ramazzini, Alessia Rianda, Gabriele Rinonapoli, Silvia Rosati, Dorotea Rubino, Martina Rueca, Alberto Ruggeri, Alessandra Sacchi, Francesco Sanasi, Carmen Santagata, Pietro Scanzano, Alessandra Scarabello, Silvana Scarcia, Vincenzo Schininà, Paola Scognamiglio, Laura Scorzolini, Eliana Specchiarello, Giulia Valeria Stazi, Giacomo Strano, Fabrizio Taglietti, Chiara Taibi, Giorgia Taloni, Tetaj Nardi, Maria Virginia Tomassi, Roberto Tonnarini, Simone Topino, Martina Tozzi, Francesco Vaia, Francesco Vairo, Maria Beatrice Valli, Alessandra Vergori, Alberta Villanacci, Laura Vincenzi, Ubaldo Visco-Comandini, Serena Vita, Pietro Vittozzi, Antonella Zanetti and Sara Zito. Open access funding provided by BIBLIOSAN. The study was performed in the framework of the SARS-CoV-2 surveillance and response program implemented by the Lazio Region Health Authority. This study was supported by funds to the National Institute for Infectious Diseases “Lazzaro Spallanzani,” IRCCS, Rome (Italy), from Italian Ministry of Health (Programme CCM 2020; Ricerca Corrente—Linea 1 on emerging and re-emerging infections); the European Commission—Horizon 2020 CoNVat, Grant agreement ID 101003544; Innovative Medicines Initiative 2 Joint Undertaking (JU) with support from European Union's Horizon 2020 research and innovation programme and EFPIA (EU project 101005075-KRONO); SHARP, Grant Number 848096.
ACKNOWLEDGMENTS
Open access funding provided by BIBLIOSAN.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The study was approved by the Ethical Committee of the National Institute for Infectious Diseases “Lazzaro Spallanzani” in Rome, Italy, as National Review Board for COVID-19 pandemic in Italy. All procedures contributing to the work described comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. All the participants signed the informed consent before entering the study.
Open Research
DATA AVAILABILITY STATEMENT
The raw data generated and/or analyzed within the present study are available in our institutional repository (rawdata.inmi.it), subject to registration. In the event of a malfunction of the application, the request can be sent directly by e-mail to the Library ([email protected]). No charge for granting access to data is required.