2022 outbreak of human mpox: Dissemination and evolution dynamic
Hanlin Liu and Zifeng Pang contributed equally to this work.
Mpox (monkeypox), which is similar to smallpox, was first reported in Central Africa in the 1970s and was once predominantly endemic in East and Central African countries.1 Since May 6, 2022, there has been a dramatic increase in confirmed cases of monkeypox worldwide. Today, the number of confirmed monkeypox cases continues to climb, causing sustained and widespread concern worldwide. We have been monitoring the prevalence of mpox virus (MPXV) and systematically analyzing its development in recent months from the perspective of dissemination and genetic evolution dynamics.
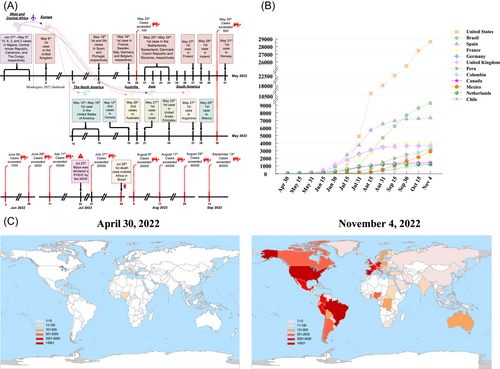
On May 6, 2022, the first case of the disease outside nonendemic areas was confirmed in the United Kingdom in a male returning from Nigeria. As of May 20, cases of mpox had been reported in 12 nonendemic countries, and the total number of infections has exceeded 100 (Figure 1A). MPXV was found in some cases that had no established travel links to the endemic areas of West and Central Africa (Table S1). The ongoing and unprecedented community transmission of MPXV has been confirmed among gay, bisexual, and other men who have sex with men seen in many nonendemic countries. As of 21 days after the first confirmed case in Europe, cases of mpox were reported on every continent except Antarctica (Figure 1A). Mpox was declared a public health emergency of international concern by the World Health Organization on July 23 (Figure 1A).2 On July 29, Brazil reported its first death outside of Africa, when a 41-year-old man being treated for lymphoma died of sepsis.3 The spread of the 2022 MPXV outbreak from January to September is shown in Appendix S1. On September 13, the total number of confirmed cases exceeded 60 000 (Figure 1A). As of November 2022, mpox cases were reported in more than 100 countries and territories around the world (Figure 1B,C). The cumulative number of confirmed cases exceeded 1000 in 12 countries worldwide, all of which were nonendemic countries located in Europe and the Americas (Figure 1B,C). The rapid spread of mpox from endemic to nonendemic countries is a worrying phenomenon, suggesting that any potential infection can spread worldwide in a short time. These dissemination events also remind us that natural geographic barriers against pathogens can now be easily broken through by trade and travel.
We retrieved all 827 MPXV genomes from GISAID as of July 30, 2022. The phylogenetic tree showed that MPXVs were divided into three clades (clades 1, 2, and 3), which were assigned in accordance with Wang's research4 (Figure 2A). hMPXV-1A in clade 3 was further divided into lineages A, A.1, A.1.1, and B.1 for the 2022 MPXV global outbreak. Lineage B.1 (n = 607) includes all MPXV genomes for the 2022 outbreak (Table S2). Four months later, we obtained the ML tree from nextstrain, which contained 1122 MPXV genomes sampled from October 2017 to November 2022.5 The total number had increased to 23 lineages (Figure 2B and Table S3). All lineages in clade B were first isolated in nonendemic countries after May 2022 (Figure 2C). B.1 has been the predominant lineage in nonendemic countries thus far. There were significant differences in the lineage characteristics of MPXV between endemic and nonendemic countries. The predominant lineages in endemic countries were A and A.1‒A.3, while those in nonendemic countries were B and B.1.1‒B.1.14. High lineage diversity was also found in the 11 representative nonendemic countries, including the United States, the United Kingdom, Germany, and Colombia (Figure 2D).
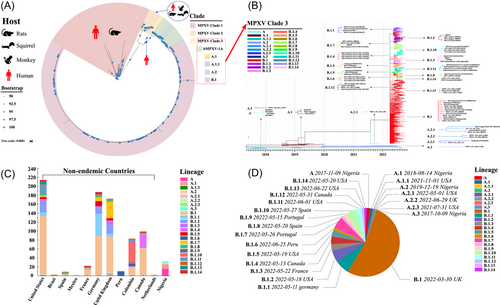
The rapidly spreading mpox epidemic and emergence of novel taxonomic lineages in the phylogenetic tree have reflected the accelerated evolution of human mpox. This suggests that MPXV may have undergone a new adaptive evolution during its global dissemination. This adaptive evolution is distinctly geographically diverse, and there may be multiple factors responsible for this diversity, such as frequent trade and travel in developed Western countries and the influence of climatic factors in different regions. In addition, it is unclear whether this evolution is affected by repeated cross-infection between humans, companion animals, and wildlife, which are issues that should be researched. The current rapid spread and evolution of mpox is unprecedented and poses a continuing threat to public health. Controlling its spread and prevalence is also a primary challenge.
AUTHOR CONTRIBUTIONS
Hanlin Liu: conceptualization; methodology; data curation; formal analysis; writing—original draft; writing—review and editing. Zifeng Pang: data curation; resources; investigation. Quan Liu: formal analysis; investigation. Hailiang Sun: writing—review and editing. Ming Liao: formal analysis; writing—review and editing.
ACKNOWLEDGMENTS
We acknowledge the authors and the originating and submitting laboratories of the sequences from GISAID's Monkeypox Database on which this research is based. All submitters of data can be contacted directly through the GISAID website (https://www.gisaid.org). This work was supported by special fund for scientific innovation strategy-construction of high-level Academy of Agriculture Science-Distinguished Scholar (R2020PY-JC001).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
DATA AVAILABILITY STATEMENT
Sequences used in this study were retrieved from the GISAID and Genbank database, and their corresponding accession numbers are listed in Supplementary Table S2.




