The natural killer cell-associated rs9916629-C allele is a novel genetic risk factor for fatal COVID-19
Abstract
The severity of COVID-19 is associated with individual genetic host factors. Among these, genetic polymorphisms affecting natural killer (NK) cell responses, as variations in the HLA-E- (HLA-E*0101/0103), FcγRIIIa- (FcγRIIIa-158-F/V), and NKG2C- (KLRC2wt/del) receptor, were associated with severe COVID-19. Recently, the rs9916629-C/T genetic polymorphism was identified that indirectly shape the human NK cell repertoire towards highly pro-inflammatory CD56bright NK cells. We investigated whether the rs9916629-C/T variants alone and in comparison to the other risk factors are associated with a fatal course of COVID-19. We included 1042 hospitalized surviving and 159 nonsurviving COVID-19 patients as well as 1000 healthy controls. rs9916629-C/T variants were genotyped by TaqMan assays and were compared between the groups. The patients' age, comorbidities, HLA-E*0101/0103, FcγRIIIa-158-F/V, and KLRC2wt/del variants were also determined. The presence of the rs9916629-C allele was a risk factor for severe and fatal COVID-19 (p < 0.0001), independent of the patients' age or comorbidities. Fatal COVID-19 was more frequent in younger patients (<69.85 years) carrying the FcγRIIIa-158-V/V (p < 0.006) and in older patients expressing the KLRC2del variant (p < 0.003). Thus, patients with the rs9916629-C allele have a significantly increased risk for fatal COVID-19 and identification of the genetic variants may be used as prognostic marker for hospitalized COVID-19 patients.
1 INTRODUCTION
Since the first emergence in 2019, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global, ongoing pandemic. While the majority of SARS-CoV-2 infections result in mild symptoms, some patients experience a severe or even fatal course of coronavirus disease 2019 (COVID-19). Different risk factors, such as the infecting SARS-CoV-2 variant, the patients' age and sex as well as chronic pulmonary, cardiovascular, kidney, neurological, malignant, or metabolic diseases were associated with progression to severe COVID-19.1 It was, however, not fully clarified, why some COVID-19 patients decease, while others recover from severe infections. An increasing number of studies indicates that distinct host genetic factors substantially contribute to the progression to fatal COVID-19.2 Importantly, among the host genes, which were recently associated with severe COVID-19, the majority were associated with CD56bright natural killer (NK) cell responses.3
NK cells are potent pro-inflammatory lymphocytes, which play a controversial role in COVID-19 patients. On one hand, NK cells may limit the SARS-CoV-2 replication and severe infections, but on the other hand, they were associated with the aggravation of COVID-19 due to the high-level secretion of pro-inflammatory cytokines.4, 5 Distinct NK cell-associated genetic variants may have an impact on the severity of COVID-19.6 NKG2C+ NK cell responses may limit the viral dissemination and individuals encoding for heterozygous and homozygous deletions of the NKG2C-encoding KLRC2 gene (KLRC2del) may have impaired NK cell immune responses. In European populations, the NKG2C receptor, HLA-E, occurs as the high expressing HLA-E*0103 or as the low-expressing HLA-E*0101 allele, which was recently associated with impaired NKG2C+ NK cell responses and severe COVID-19.7
Another NK cell associated factor, which contributes to the severity of COVID-19, is the FCGR3A-158-V/F polymorphism in the human CD16a/FcγRIIIa antibody-receptor. Expression of the high-affinity FCGR3A-158-V/V genotype results in a potent antibody-dependent activation of CD16+CD56bright NK cells and was recently associated with severe COVID-19.8
Recently, it has been described that the rs9916629-C/T polymorphism, which is located in the untranslated region (UTR) between the human SLFN12 and SLFN13 genes, has a significant impact on the NK cell repertoire in the human host. Individuals expressing the rs9916629-C allele exhibit significantly higher CD56bright NK cell frequencies, compared to individuals expressing the rs9916629-T/T genotype.9 However, the impact of this variant on COVID-19 disease severity has so far not been investigated. Using a genetic-association study, we now investigated whether this NK cell associated genetic risk factor alone, or in combination with other risk factors, is associated with fatal outcome in hospitalized COVID-19 patients.
2 MATERIALS AND METHODS
2.1 Study cohort
In total 1201 Austrian COVID-19 patients (40.8% female, median age: 61.42), hospitalized between 02/27/2020 and 02/22/2022 in the Clinic Favoriten Vienna or the Vienna General Hospital, were included in the study. All were infected with either the SARS-CoV-2 wild-type European-lineage (D614G), or the Alpha (B.1.1.7) or Delta (B.1.617.2) variant of concern (VoC) as confirmed by specific PCR assays. All patients were hospitalized due to severe COVID-19 related symptoms (https://www.covid19treatmentguidelines.nih.gov) and were SARS-CoV-2 IgG seronegative at the onset of hospitalization. As controls, we included 1000 healthy SARS-CoV-2 PCR-negative individuals (51.1% female, median age 57.9 years) without any symptoms suggestive for COVID-19, whose plasma samples were obtained for routine vaccination titer controls. Details of the study cohort are shown in Table 1.
| Study cohort | p Valuea | ||
|---|---|---|---|
| Hospitalized survivors N = 1042 | Hospitalized deceased N = 159 | ||
| Sex | |||
| Female (%) | N = 436 (41.8%) | N = 54 (34%) | p = 0.07 |
| Male (%) | N = 606 (58.2%) | N = 105 (66%) | |
| Median age (min–max) | 58.4 (18.1–100.1) | 77.1 (48.3–100.9) | p < 0.0001 |
| Comorbidities | |||
| Nonasthmatic COPD (%) | N = 107 (10.3%) | N = 35 (22.1%) | p < 0.0001 |
| Cancer (%) | N = 140 75 (13.4%) | N = 35 (22.1%) | |
| Dementia (%) | N = 75 (7.2%) | N = 16 (9.8%) | |
| Diabetes mellitus (%) | N = 247 (23.7%) | N = 39 (24.6%) | |
| Hypertension (%) | N = 569 (54.6%) | N = 97 (61.3%) | |
| CAD (%) | N = 129 (12.4%) | N = 51 (31.9%) | |
| Chronic kidney diseases (%) | N = 140 (13.4%) | N = 39 (24.5%) | |
| Chronic liver diseases (%) | N = 64 (6.2%) | N = 8 (4.9%) | |
| Chronic neurological diseases (%) | N = 75 (7.2%) | N = 31 (19.6%) | |
| Obesity (BMI > 30 kg/m2) (%) | N = 140 (13.4%) | N = 19 (12.3%) | |
| Number of comorbidities | |||
| 0 (%) | N = 321 (30.8%) | N = 3 (1.9%) | p < 0.0001 |
| 1 (%) | N = 282 (27.0%) | N = 36 (22.6%) | |
| 2 (%) | N = 221 (21.2%) | N = 44 (27.7%) | |
| 3 (%) | N = 97 (9.3%) | N = 50 (31.4%) | |
| 4 (%) | N = 86 (8.3%) | N = 12 (7.5%) | |
| 5 (%) | N = 35 (3.3%) | N = 9 (5.6%) | |
| 6 (%) | N = 0 (0%) | N = 6 (3.8%) | |
| SARS-CoV-2 variant | |||
| WT D614G | N = 631 (60.6%) | N = 102 (64.2%) | p = 0.61 |
| Alpha (B.1.1.7) | N = 165 (15.8%) | N = 21 (13.2%) | |
| Delta (B.1.617.2) | N = 246 (23.6%) | N = 36 (22.6%) | |
- Abbreviations: BMI, body mass index; CAD, coronary artery disease; COPD, chronic obstructive pulmonary disease; WT, wild-type.
- a Differences were assessed with the χ2 (sex, number of comorbidities, and SARS-CoV-2 variant distribution) and Mann–Whitney-test (age).
2.2 SARS-CoV-2 detection and serology
Viral RNA was isolated from respiratory swabs using NucliSens EasyMag extractor (BioMérieux) and tested for SARS-CoV-2 by a recently published qPCR.10 The infecting SARS-CoV-2 Alpha- and Delta-VoCs were genotyped with VirSNiP Mutation Assays Kits according to the manufacture's instruction (TIB Molbiol). SARS-CoV-2 S1-domain-specific IgG antibodies were detected and quantified by ELISA (Euroimmune).
2.3 Genotyping
Genomic DNA was isolated from respiratory swabs or from plasma samples using NucliSens EasyMag extractor. The KLRC2wt/del, FcγRIIIa-158-F/V and HLA-E*0101/0103 variants were genotyped by touchdown PCR or TaqMan assays, respectively, according to recently published protocols.11, 12 The rs9916629-C/T variants were determined by an in-house TaqMan assay, using rs9916629-C/T-specific probes (GCATTATTAATGTACTAGTTC-VIC and GCATTATTAATGTATTAGTTC-6FAM), primers (Fwd.: CCCTCAAGCTTTCCCCCGTG and Rev: TGCAGTCAGGTTGTTGGGGG) and the Luna Universal Probe qPCR Master Mix (NEB Biolabs), according to the manufacturer's instruction.
2.4 Statistical analysis
χ2 test or Fisher's exact test were used to compare the distribution of sexes and genetic variants. Mann–Whitney-test was used to compare the age and the number of comorbidities. The age and number of comorbidities associated with a fatal outcome in hospitalized COVID-19 patients were analyzed by receiver operating characteristic (ROC) curves. A p < 0.05 was considered statistically significant. Statistical differences were assessed with GraphPad Prism 9.
3 RESULTS
3.1 rs9916629-C/T variants in hospitalized surviving and deceased COVID-19 patients
From the 1201 hospitalized COVID-19 patients included in the study, 159 (13.2%) died during hospitalization, while 1042 (86.8%) survived and recovered (Supporting Information: Figure S1). Previously known risk factors as age, sex, and comorbidities as well as the variant of the infecting SARS-CoV-2 strain, were compared between both groups. Deceased COVID-19 patients were significantly older and had more comorbidities, compared to survivors, while the patients' sex and infecting SARS-CoV-2 variant were not distributed differently between the groups (Table 1).
We then investigated the newly described rs9916629-C/T polymorphism in the 1201 hospitalized patients and in the 1000 healthy controls. As shown in Figure 1A, 8.3% of the controls, encoded for the rs9916629-C/C, while 38.2% and 53.5% of the controls showed the rs9916629-C/T and rs9916629-T/T genotype, respectively. In hospitalized survivors and even more in deceased COVID-19 patients, the rs9916629-C/C and the in rs9916629-C/T genotype were significantly over-represented, while the rs9916629-T/T only rarely occurred (Figure 1A). Thus, the rs9916629-C allele is associated with severe and fatal COVID-19 (Figure 1B).
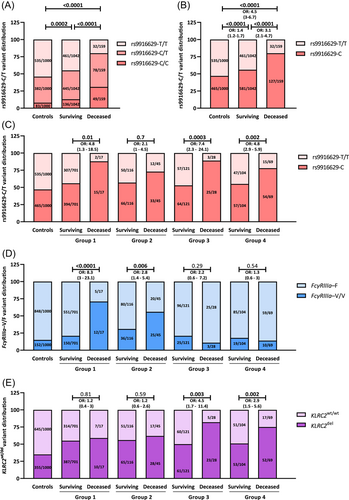
3.2 Age- and comorbidity-adjusted risk for fatal COVID-19
We then analyzed the rs9916629-C/T variants in relation to the patients' age and the number of their comorbidities. To determine the age and the number of comorbidities that are associated with an overall increased risk for fatal COVID-19, ROC analyses were performed and as shown in Supporting Information: Figures S2 and S3, an age of ≥69.85 years and the presence of ≥3 comorbidities were associated with a significantly increased risk for fatal COVID-19. We thus stratified our study cohorts into patients age <69.85 years and <3 comorbidities (N = 718, 59.8%, Group 1); patients <69.85 years and ≥3 comorbidities (N = 161, 13.4%, Group 2); patients ≥69.85 years and <3 comorbidities (N = 149, 12.4%, Group 3) as well as patients ≥69.85 years and ≥3 comorbidities (N = 173, 14.4%, Group 4).
We then compared the rs9916629-C/T variants in surviving and deceased patients in all four groups. The rs9916629-C allele was significantly overrepresented in deceased COVID-19 patients of all cohorts, except for Group 3 (Figure 1C).
3.3 FcγRIIIa-158-V/V, KLRC2wt/del, and HLA-E*0101/0103 variants in surviving or deceased COVID-19 patients
We then assessed the distribution of FcγRIIIa-158-V/V, KLRC2wt/del, and HLA-E*0101/0103 variants between controls, survivors, and deceased COVID-19 patients. Hospitalized survivors and especially deceased COVID-19 patients encoded significantly more often for the FcγRIIIa-158-V/V, and the KLRC2wt/del and KLRC2del/del variants, while in the controls the FcγRIIIa-158-F/F and KLRC2wt/wt genotypes were more prevalent (Supporting Information: Table S1). Thus, the FcγRIIIa-158-V/V genotype and the KLRC2del allele were risk factors for fatal COVID-19 (Table 2). Although the HLA-E*0101 allele was overrepresented in hospitalized COVID-19 patients in general (p = 0.049, OR:1.2 (95% CI: 1–1.4), no significant different variant distribution was found between deceased and surviving COVID-19 patients (Table 2).
| Study cohort | p Valuea | ||
|---|---|---|---|
| Hospitalized survivors N = 1042 | Hospitalized deceased N = 159 | ||
| FcγRIIIa risk genotype | |||
| FcγRIIIa–V/V | N = 230 (22.1%) | N = 50 (31.4%) | p = 0.01 OR: 1.6 (1.1–2.3) |
| FcγRIIIa protective allele | |||
| FcγRIIIa–F | N = 812 (77.9%) | N = 109 (68.6%) | |
| KLRC2 risk allele | |||
| KLRC2del | N = 566 (54.3%) | N = 113 (71.1%) | p < 0.0001 OR: 2.1 (1.4–2.9) |
| KLRC2 protective genotype | |||
| KLRC2wt/wt | N = 476 (45.7%) | N = 46 (28.9%) | |
| HLA-E risk allele | |||
| HLA-E*0101 | N = 814 (78.1%) | N = 123 (77.4%) | p = 0.83 OR: 1.1 (0.7–1.6) |
| HLA-E protective genotype | |||
| HLA-E*0103/0103 | N = 228 (21.9%) | N = 36 (22.6%) | |
- Abbreviations: Del, deletion; OR, odds ratio (95% confidence interval); WT, wild-type.
- a Differences were assessed with the χ2-test.
We then compared the frequency of the FcγRIIIa-158-V/F and KLRC2wt/del variants in all four groups defined by age and comorbidity. As shown in Figure 1D,E, THE FcγRIIIa-158-V/V genotype was significantly overrepresented only in deceased COVID-19 patients <69.85 years (Group 1 and 2, Figure 1D), while the KLRC2del allele was significantly overrepresented solely in deceased older patients (Group 3 and 4, Figure 1E). The distribution of the HLA-E*0101/0103 variants did not differ between groups (Supporting Information: Figure S4).
3.4 Combination of risk factors in surviving or deceased COVID-19 patients
We next analyzed whether a specific combination of genetic risk factors is associated with an increased risk for fatal COVID-19. The combination of both risk factors, rs9916629-C, and FcγRIIIa-158-V/V, dominated in deceased younger and occurred rarely in surviving COVID-19 patients (Figure 2A, Group 1: 70.6% and 6.7%, OR: 33.4 (95% CI: 11.3–98.8); Group 2: 48.9% and 20.7%, OR:3.7 (95% CI: 1.8–7.7). Surviving patients with ≥3 comorbidities encoded significantly more frequently for both protective factors (rs9916629-T/T and FcγRIIIa-158-F), compared to surviving patients with <3 comorbidities.

In both groups of older patients, the combination of both risk factors, rs9916629-C and KLRC2del, was significantly overrepresented in deceased COVID-19 patients (Figure 2B, Group 3: 71.4% and 26.4%, OR:6.9 (95% CI: 2.8–17.4); Group 4: 58% and 26.9%, OR:3.7 (95% CI: 2–7.1).
4 DISCUSSION
In this study, we identified the rs9916629-C/T polymorphism as a novel genetic risk factor for fatal COVID-19. Although the patients' age and distinct comorbidities are known risk factors for severe COVID-19,1 these factors alone cannot predict which hospitalized COVID-19 patients have a high risk to decease. We now demonstrate that the presence of the rs9916629-C allele is associated with a significantly increased risk for fatal COVID-19. The rs9916629-C/T polymorphism has only recently been identified as a major factor influencing the human NK cell repertoire, and both, the rs9916629-C/C and rs9916629-C/T variants are associated with increased CD56bright NK cell frequencies, potentially via increased RNA expression levels of the transcription factor TBX21.9
CD56bright NK cells are a NK cell subset, which releases high-levels of pro-inflammatory cytokines.6 A recently published genome-wide association study localized the genetic risk for fatal COVID-19 in particular to CD56bright NK cells.3 Additional ex vivo studies demonstrated that arming and activation of CD56bright cells occurs especially in severely diseased COVID-19 patients.13, 14 Other studies demonstrated that the CD56bright-derived IFN-γ, TNF-α, GM-CSF and IL-13 levels are highly elevated in the serum of severely ill COVID-19 patients, and may contribute to the disadvantageous immunopathogenesis, leading to a severe course of COVID-19.15-18 This supports our data, showing that the rs9916629 polymorphism is highly associated with fatal disease, as the rs9916629-C allele leads to high CD56bright frequencies, which potentially exaggerate the release of pro-inflammatory cytokines.
Between 30% and 50% of all CD56bright NK cells express the CD16a/FcγRIIIa-receptor6 and we recently identified the high-affinity FCGR3A-158-V/V genotype as a risk factor for severe COVID-19.19 In the present study, we could confirm this finding. In addition, we were also able to further specify that the FCGR3A-158-V/V variant was highly associated with an increased risk for a fatal disease only in younger COVID-19 patients and especially in individuals, who also encoded for the rs9916629-C allele. The presence of both, the rs9916629-C and FcγRIIIa-158-V/V variant could result in high CD56brightCD16+ NK cell levels, which are further highly activated by the high-affinity FcγRIIIa-158-V/V variant and thus contribute to exaggerated NK cell-mediated pro-inflammatory cytokine responses.
In our study cohort, also the KLRC2del allele was associated with an increased risk for a fatal outcome in COVID-19 patients. The data from our large study cohort are in agreement with previously published studies, which identified the KLRC2wt/del and KLRCdel/del genotypes as independent risk factors for severe COVID-19 and demonstrated that high NKG2C+ NK cell counts are frequently found in COVID-19 survivors.12, 20 This underlines the important role of potent antiviral NKG2C+ NK cell responses in the prevention of severe COVID-19. The combination of the KLRC2del and the rs9916629-C allele was associated with a particularly high mortality in older COVID-19 patients, independent from the patients' number of comorbidities. So far, it is unknown whether and to which extent the rs9916629-C/T polymorphism affects the frequency and function of NKG2C+ NK cells. Further ex vivo and in vitro studies are thus required to analyze the impact of the rs9916629-C/T polymorphism on the NKG2C+ NK cell repertoire, the functional SARS-CoV-2 specific NK cell responses as well as the COVID-19 pathogenesis.
In conclusion, we demonstrate that the rs9916629-C/T polymorphism contributes significantly to the risk of fatal COVID-19. The analysis of the rs9916629-C/T variants and especially together with FCGR3A-158-V/V and KLRC2del may help to identify younger and older hospitalized patients at risk for fatal COVID-19, respectively. Overall, our data are of special interest as predictive markers for fatal COVID-19 in hospitalized COVID-19 patients are still scarce. However, further and extended studies are needed to confirm whether the analysis of NK cell-associated genetic risk factors may be used as a predictive marker for fatal COVID-19.
AUTHOR CONTRIBUTIONS
Hannes Vietzen: Conceptualization; data curation; formal analysis; investigation; methodology; validation; visualization; writing – original draft. Elisabeth Puchhammer-Stöckl: Conceptualization; funding acquisition; investigation; project administration; supervision; writing – original draft. Philippe L. Furlano: Data curation; formal analysis; investigation; methodology. Marianna Traugott: Resources. David Totschnig: Resources. Wolfgang Hoepler: Resources. Robert Strassl: Resources. Alexander Zoufaly: Resources.
ACKNOWLEDGMENT
The study was funded by the Center for Virology, Medical University of Vienna, Vienna, Austria.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The study was approved by the institutional review board of the Medical University of Vienna (EK No. 1143/2022).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data are not publicly available due to privacy or ethical restrictions.




