The emergence of recombinant norovirus GII.12[P16] and predominance of GII.3[P12] strains in pediatric patients with acute gastroenteritis in Thailand, 2019–2020
Abstract
Norovirus (NoV) and sapovirus (SaV) are important pathogens that cause acute gastroenteritis (AGE) in all age groups, commonly in children worldwide. Recently, a number of studies have reported a wide variety of NoV recombinant strains. This study aimed to investigate the distribution of NoV and SaV recombinant strains circulating in Chiang Mai, Thailand, during 2019–2020. One hundred and twenty-four NoV and seven SaV strains detected in children admitted to the hospital with AGE were included in this study. The partial RNA-dependent RNA-polymerase (RdRp)/VP1 regions of these NoV and SaV strains were analyzed by phylogenetic analysis, Simplot, and RDP software. Overall, eight recombination patterns of NoV were detected. NoV GII.4[P16] was the most common strain detected (39.1%), followed by GII.3[P12] (25.0%), GII.4[P31] (17.2%), and other recombinant strains were detected at a lower rate. NoV GII.12[P16] strains were detected for the first time in Thailand. For SaV, none of the recombinant strains was detected. All SaV strains, GI.1/GI.1, GI.2/GI.2, and GII.5/GII.5, exhibited VP1 genotype corresponded to RdRp genotype. In conclusion, this study demonstrates the distribution and diversity of NoV and SaV recombinant strains circulating in pediatric patients with AGE in Chiang Mai, during 2019–2020 with the emergence of NoV GII.3[P12] and GII.12[P16].
1 INTRODUCTION
Noroviruses (NoVs) are recognized as one of the most common nonbacterial agents to cause acute gastroenteritis (AGE) in all age groups, especially in children under 5 years of age worldwide. Approximately 200 000 deaths and $60 billion of financial cost per year are attributable to NoV infection globally.1, 2 Moreover, the prevalence of NoV infection tends to increase after the implementation of the rotavirus vaccine and leads to a major burden of AGE.3, 4 Sapovirus (SaV), a member in the same family with NoV, can also cause AGE with less severity than NoV infection. Nevertheless, the mortality and increasing hospitalization caused by SaV infection has been demonstrated.5
On the basis of the amino acid sequence of major capsid protein (VP1), NoV is currently classified into 10 genogroups (GI-GX). These genogroups can be subdivided into 48 confirmed genotypes. NoV GI, GII, GIV, GVIII, and GIX can infect and cause illness in humans.6 Among these, 37 genotypes of NoV detected in the human population have been reported worldwide.7 NoV GII.4 predominate continuously over 2 decades with six major pandemic variants have been identified.8 On the basis of partial RNA-dependent RNA-polymerase (RdRp) nucleotide (nt) sequences, NoV is divided into eight polymerase (P) groups (GI.P-GVII.P and GX.P). Each P-group is further subdivided into a total of 60 P-types.6 To date, 56 P-types have been identified in humans. NoV GII.P4 is the most predominant P-type. However, the emergence of other P-types such as GII.P16, GII.P17, and GII.P31 have been documented and recently became the most common P-types during 2013-2019.7
Since the recombination event of NoV has been increasingly reported worldwide, dual-typing (genotype and P-type) is recently proposed for the nomenclature of NoV strain. So far, more than 30 recombinant patterns have been reported from 2015 to 2020. Of these, NoV GII.4 Sydney [P16] and GII.4 Sydney [P31] are the most predominant in many countries.9 At the same time, an increase of other recombinant NoV such as GII.2[P16] and GII.3[P12] as the predominant type has been demonstrated in Malaysia and Germany.10, 11 In addition, the emergence of NoV nonrecombinant strain GII.17[P17] has also been reported during 2013–2015 in Japan and USA.12, 13
SaV is divided into 19 genogroups (GI to GXIX) based on amino acid sequences of the VP1 region. SaV GI, GII, GIV, and GV infect humans14, 15 and SaV GI.1 is the predominant genotype. Nevertheless, the study on SaV recombination is still limited.16, 17 In Chiang Mai, Thailand, investigation of NoV and SaV recombinant strains has been continually carried out from 2005 to 2018 and the data demonstrated that NoV recombinant strains circulating in this area have changed over time throughout the study period.18-20 This study aimed to investigate the distribution and diversity of NoV and SaV recombinant strains circulating in pediatric patients hospitalized with AGE in Chiang Mai, during 2019–2020.
2 MATERIALS AND METHODS
2.1 Stool sample collection
Stool samples were collected from children less than 5 years of age who were admitted to five major hospitals in Chiang Mai, including Chiang Mai University Hospital, Nakornping Hospital, Rajavej Hospital, Sanpatong Hospital, and Sriphat Medical Center during a period of 2 years from January 2019 to December 2020. The inclusion criteria for the collection of fecal specimens were inpatients with AGE who presented loose or watery diarrhea more than three times within 24 h. All stool samples were stored at −20°C until use. The study was conducted with the approval of the Institutional Review Board of the Faculty of Medicine, Chiang Mai University (MIC-2557-02710).
2.2 Extraction of the viral genome, reverse transcription, and detection of NoV and SaV
Viral genomic RNA was extracted from 10% stool suspension using a Viral Nucleic Acid Extraction Kit (Geneaid) according to manufacturer's instructions. The extracted RNA was reverse transcribed to complementary DNA (cDNA) using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Screening of NoV and SaV was performed by using multiplex-polymerase chain reaction (PCR) and seminested PCR as described previously21, 22 using GoTaq DNA Polymerase (Promega).
2.3 Amplification and sequencing of partial RdRp/VP1 region of NoV GII and SaV
The partial RdRp/VP1 region of NoV GII positive samples was amplified by seminested PCR. For the first round PCR, a 2 µl of cDNA was added into 23 µl of PCR mixture containing primers JV12Y23 and G2SKR. The first PCR products were further subjected to the second PCR amplification using primers P290mo24 and G2SKR. The expected sizes of PCR products from the first and second round amplifications were 1111 and 1095 bp, respectively. Additionally, another set of primers, CB0125/G2SKR and NV461126/G2SKR, was used for the amplification of the samples that could not be amplified by the first set of primers. The expected sizes of PCR products in the first and second round amplifications were 1146 and 1052 bp, respectively. For SaV, positive samples were amplified using primers Sapp3627/SaV1245R28 and Sapp12829/SaV1245R28 for the first and second round PCR with the amplicons size of 905 and 824 bp, respectively. All PCR products were purified using GenepHlow Gel/PCR Kit (Geneaid) according to the manufacturer's protocol. The nt sequencing of purified PCR products was performed by first BASE Laboratories (Apical Science).
2.4 Analysis of NoV and SaV recombinants
The obtained nt sequences of NoV and SaV were analyzed to identify P-type and genotype using BLAST server (http://blast.ncbi.nlm.nih.gov), Norovirus Genotyping Tool Version 2.0 (https://www.rivm.nl/mpf/typingtool/norovirus/), and Human Calicivirus Typing Tool (https://calicivirustypingtool.cdc.gov). To confirm genotype, P-type, and recombination event of NoV and SaV strains, phylogenetic trees of RdRp and VP1 genes were constructed using Mega11 software30 with the maximum-likelihood method. The best fit DNA/protein model was carried out for the method setting. Simplot and RDP4 software (Version 4.39) were used to evaluate and predict the potential recombination breakpoint with statistically significant analysis. Lordsdale norovirus (accession no. X86557) was used as the reference to locate the nt position breakpoint on NoV genome. NoV GII prototype strains including GII.2[P2] (X81879), GII.3[P3] (U02030), GII.4[P4] (X76716 and GU445325), GII.4[P31] (AB434770), GII.6[P6] (AB039778), GII.7[P7] (AB231342), GII.12[P12] (AB039775), and GII.16[P16] (AY772730) were used for recombination analysis.
2.5 The nt sequence accession numbers of NoV and SaV
nt sequences of partial RdRp/VP1 region of NoV and SaV strains analyzed in this study are available in the GenBank database under the accession numbers OP349114-OP349177and OP352888-OP352894.
3 RESULTS
3.1 Genotype and P-type distribution of NoV and SaV
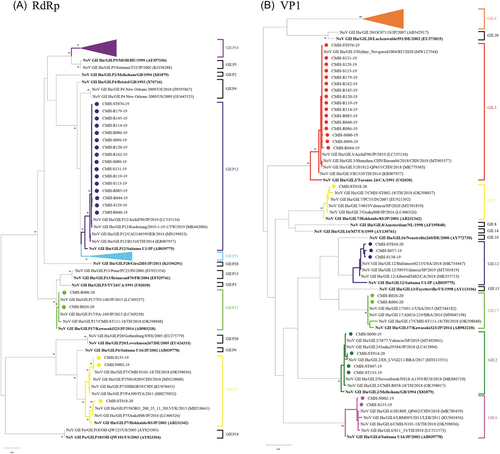
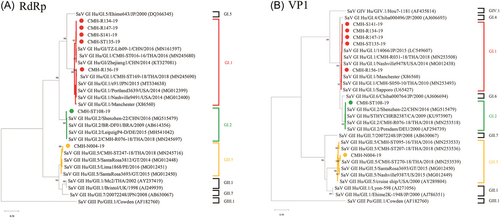
From a total of 124 NoV GII positive samples, 64 were successfully amplified for the partial RdRp/VP1 region. Of these, 51 and 13 samples were collected in 2019 and 2020, respectively. Genotype (VP1 gene) of NoV strains detected in this study included GII.4 (36 strains), GII.3 (16 strains), GII.2 (4 strains), GII.12 (3 strains), GII.6 (2 strains), GII.17 (2 strains), and GII.7 (1 strain). Meanwhile, the P-type (RdRp gene) of these strains was also analyzed. It was found that 32, 16, 11, 3, and 2 strains were identified as NoV GII.P16, GII.12, GII.P31, GII.P7, and GII.P17, respectively. For SaV, all seven positive samples could be amplified for partial RdRp/VP1 gene. Analysis of the capsid VP1 gene, three genotypes of SaV were identified including GI.1 (five strains), GI.2 (one strain), and GII.5 (one strain). Similarly, the RdRp genotype of each SaV strain was also analyzed and found to be corresponded with its genotype of the VP1 gene, suggesting that all SaV strains were nonrecombinant strains (Table 1).
| Genotype and P-type | No. of positive cases (%) | Total cases (%) | |
|---|---|---|---|
| 2019 | 2020 | ||
| NoV | 51 | 13 | 64 |
| GII.4 Sydney 2012[P16] | 17 (33.3) | 8 (61.5) | 25 (39.1) |
| GII.3[P12] | 16 (31.4) | 0 | 16 (25.0) |
| GII.4 Sydney 2012[P31] | 11 (21.6) | 0 | 11 (17.2) |
| GII.2[P16] | 3 (5.9) | 1 (7.7) | 4 (6.3) |
| GII.12[P16] | 2 (3.9) | 1 (7.7) | 3 (4.7) |
| GII.6[P7] | 2 (3.9) | 0 | 2 (3.1) |
| GII.17[P17] | 0 | 2 (15.4) | 2 (3.1) |
| GII.7[P7] | 0 | 1 (7.7) | 1 (1.6) |
| SaV | 7 | 0 | 7 |
| GI.1/GI.1 | 5 (71.4) | 0 | 5 (71.4) |
| GI.2/GI.2 | 1 (14.3) | 0 | 1 (14.3) |
| GII.5/GII.5 | 1 (14.3) | 0 | 1 (14.3) |
3.2 Recombination patterns of NoV and SaV
As shown in Table 1, six recombinant strains including NoV GII.2[P16], GII.3[P12], GII.4 Sydney 2012[P16], GII.4 Sydney 2012[P31], GII.6[P7], GII.12[P16], and two nonrecombinant strains GII.7[P7] and GII.17[P17] were detected in this study. NoV GII.4 Sydney 2012[P16] was the most predominant (39.1%) followed by GII.3[P12] (25.0%), and GII.4 Sydney 2012[P31] (17.2%) while other recombinant strains were detected with lower prevalence. The genotype and P-type of NoV strains detected in this study were confirmed by phylogenetic analysis of VP1 and RdRp genes, respectively (Figure 1). In this study, the SaV recombinant strain was not detected. All SaV strains detected in this study, GI.1/GI.1, GI.2/GI.2, and GII.5/GII.5 exhibited VP1 genotypes corresponding to their RdRp genotypes with the predominant of GI.1/GI.1 (71.4%) (Table 1). Phylogenetic trees of partial RdRp and VP1 genes of SaV detected in this study are shown in Figure 2.
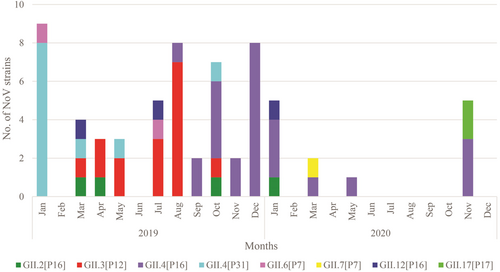
3.3 Monthly distribution of NoV recombinant and nonrecombinant strains
In this study, the monthly distribution of NoV strains circulating in children with AGE during 2019–2020 was further analyzed (Figure 3). The results revealed that different NoV strains predominated in different time periods. NoV GII.4[P31] was predominant in January 2019. NoV GII.3[P12] emerged from March to August 2019. Afterward, the NoV GII.3[P12] was replaced by NoV GII.4[P16] in September 2019. Then, NoV GII.4[P16] became the most common strain detected at the end of 2020.
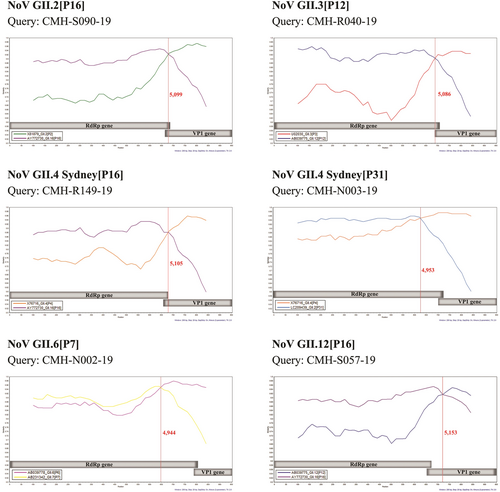
3.4 Analysis of recombination breakpoint
From a total of 64 NoV strains detected in this study, 61 strains presented their VP1 genotypes in conflict with RdRp genotypes, indicating that they were NoV recombinant strains. All NoV recombinant strains were further analyzed for recombination breakpoint using Simplot and RDP analyses. The results showed that recombination sites were located within the open reading frame (ORF)1, ORF1/2 junction, or ORF2 region between nt positions 4944 and 5153 (Table 2, Figure 4). Three NoV GII.2[P16] strains showed recombination breakpoint at nt position 5099 while another strain showed recombination breakpoint at nt position 5037. All 16 NoV GII.3[P12] strains exhibited recombination breakpoint at the same nt position at 5086. The NoV GII.4[P16] recombinant strains displayed two different recombination breakpoints at nt positions 5077 (2 strains) and 5105 (23 strains) (Table 2). In contrast, the NoV GII.4[P31] recombination strains displayed three different recombination breakpoints at nt position 4953 (nine strains), 4973 (one strain), and 5105 (one strain). The difference in the recombination breakpoint was also observed among the NoV GII.6[P7] and GII.12[P16] recombinant strains (Table 2).
| Recombination pattern | NoV reference strains used for Simplot and RDP4 analysis | No. of NoV recombinant strains | Predicted recombination breakpoint | ORF | p Value (RDP4) | |
|---|---|---|---|---|---|---|
| VP1 (accession no.) | RdRp (accession no.) | |||||
| GII.2[P16] | GII.2[P2] (X81879) | GII.16[P16] (AY772730) | 1 | 5037 | 1 | 1.78 × 10−15 |
| 3 | 5099 | 1/2 | 7.42–288.1 × 10−18 | |||
| GII.3[P12] | GII.3[P3] (U02030) | GII.12[P12] (AB039775) | 16 | 5086 | 1/2 | 6.56–41.33 × 10−16 |
| GII.4 Sydney [P16] | GII.4[P4] (X76716) | GII.16[P16] (AY772730) | 2 | 5077 | 1 | 4.23–4.55 × 10−14 |
| 23 | 5105 | 2 | 1.97–84.92 × 10−15 | |||
| GII.4 Sydney [P31] | GII.4[P4] (X76716) | GII.2[P31] (LC209439) | 9 | 4953 | 1 | 1.25–4.96 × 10−10 |
| 1 | 4973 | 1 | 1.59 × 10−13 | |||
| 1 | 5105 | 2 | 5.05 × 10−11 | |||
| GII.6[P7] | GII.6[P6] (AB039778) | GII.7[P7] (AB231342) | 1 | 4944 | 1 | 4.99 × 10−7 |
| 1 | 4971 | 1 | 2.01 × 10−7 | |||
| GII.12[P16] | GII.12[P12] (AB039775) | GII.16[P16] (AY772730) | 1 | 5151 | 2 | 4.61 × 10−7 |
| 2 | 5153 | 2 | 5.74–20.31 × 10−6 | |||
- Abbreviations: NoV, norovirus; ORF, open reading frame; RdRp, RNA-dependent RNA polymerase.
4 DISCUSSION
Recombination is one of the mechanisms that contribute a major role to the evolution and genetic diversity of NoV. Investigation of NoV recombinant strains circulating in pediatric patients with AGE in Chiang Mai has been carried out continually from 2005 to 2018.18, 19 So far, 11 different NoV recombinant patterns have been identified, including GII.1[P33], GII.2[P16], GII.3[P16], GII.3[P21], GII.4[P12] GII.4[P16], GII.4[P31], GII.6[P7], GII.12[P33], GII.13[P16], and GII.14[P7]. The emergence of NoV GII.2[P16] recombinant strains during 2016–2017 was reported in many countries worldwide as well as Thailand.11, 19, 31-33 The follow-up surveillance in Chiang Mai, from 2017 to 2018, demonstrated that the predominance of GII.2[P16] in 2017 was replaced by GII.4[P16] and GII.4[P31] in 2018.19 In this study, NoV GII.2[P16] continued to be detected both in 2019 and 2020 (Table 1). Interestingly, our data demonstrated the copredominance of three NoV recombinant strains, GII.4[P16], GII.4[P31], GII.3[P12], in 2019 and then NoV GII.3[P12] and GII.4[P31] were suddenly disappeared in 2020 whereas the NoV GII.4[P16] remained circulating as the most predominant genotype. It should be remarked that NoV GII.3[P12] and GII.12[P16] recombinant strains have never been detected previously in Chiang Mai, even though these recombination patterns have been detected previously in Bangkok, Thailand. Focusing on the P-type (RdRp gene), the NoV GII.P16 was the most predominant in this study. The GII.P16 has been demonstrated to associate with several capsid VP1 genotypes, including GII.1-GII.4, GII.10, GII.12, and GII.13.34 Molecular analysis of the RdRp region of NoV genogroup II revealed that NoV GII.P16 had the highest evolutionary rate and this might increase the possibility of recombination to occur in combination with several other VP1 genotypes.35
The emergence of a novel NoV GII.12[P16] recombinant strain was reported previously in Canada.36 Furthermore, an increase prevalence of NoV GII.3[P12] was also reported previously in Russia37 and China.38 In this study, GII.12[P16] strains were reported for the first time in Thailand, while GII.3[P12] were detected for the first time in Chiang Mai with high prevalence compared to other studies conducted in other regions of Thailand.39-41 Thus, it is interesting to keep monitoring the emergence of NoV GII.3[P12] and GII.12[P16] recombinant strains in Thailand and other geographical regions. Additionally, it should be pointed out that the recombination breakpoint of all NoV GII.3[P12] strains detected in this study were located at the same nt position (5086). Moreover, the nt sequence identities of both RdRp and VP1 genes of NoV GII.3[P12] strains were highly similar among themselves and closely related to GII.3[P12] strains detected in Bangkok in 2014.39 The data suggest that the NoV GII.3[P12] strain might be descended from the same ancestral strain, and the emergence of GII.3[P12] in this area might be disseminated and underwent recombination event from the strains circulating in Bangkok.
Detection of SaV recombinant was also performed in the current study. However, all SaV strains were nonrecombinant. Phylogenetic analysis of RdRp and VP1 genes showed that all SaV strains exhibited the same genotype of both RdRp and VP1 regions. Similar to other studies, SaV recombinant was not frequently observed or even detected at very low prevalence. We speculate that a low infection rate of SaV infection could lead to a minimal possibility for a recombination event to occur.
In this study, only 64 out of 124 (51.6%) NoV strains that their RdRp regions could be amplified and their P-types could be assigned. The limitation of this study is that approximately half of NoV strains detected in this study could not be included for identification of recombinant strain due to lacking RdRp sequence. The primer sets used in this study may fail to match with the RdRp gene of about half of NoV strains detected in this study. Therefore, designing new universal primers for amplifying the RdRp regions is required to fill the gap of NoV dual-typing in the future.
In conclusion, this study reported the distribution and diversity of NoV and SaV recombinant strains circulating in pediatric patients with AGE in Chiang Mai from 2019 to 2020 with the emergence of NoV GII.3[P12] and GII.12[P16]. The findings provide useful information about the genetic evolution of NoV recombinant strains and the predominant strains had changed over time.
ACKNOWLEDGMENTS
This research work was jointly supported by the Center of Excellence in Emerging and Re-emerging Diarrheal Viruses, Chiang Mai University, Chiang Mai, Thailand (Grant No. CoE 13/65) and Thailand Science Research and Innovation (Grant No. FF 65/026).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The study was conducted with the approval of the Institutional Review Board of the Faculty of Medicine, Chiang Mai University (MIC-2557-02710).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. The nucleotide sequences of RNA-dependent RNA polymerase and VP1 regions of norovirus and sapovirus strains described in this study have been deposited in the GenBank database under the accession numbers OP349114–OP349177 and OP352888–OP352894.




