Evaluation and validation of an RT-PCR assay for specific detection of monkeypox virus (MPXV)
Alberto Paniz-Mondolfi, and Juan David Ramírez contributed equally to this study.
Abstract
Monkeypox virus (MPXV) is a zoonotic orthopoxvirus within the Poxviridae family. MPXV is endemic to Central and West Africa. However, the world is currently witnessing an international outbreak with no clear epidemiological links to travel or animal exposure and with ever-increasing numbers of reported cases worldwide. Here, we evaluated and validated a new, sensitive, and specific real-time PCR-assay for MPXV diagnosis in humans and compare the performance of this novel assay against a Food & Drug Administration-cleared pan-Orthopox RT-PCR assay. We determined specificity, sensitivity, and analytic performance of the PKamp™ Monkeypox Virus RT-PCR assay targeting the viral F3L-gene. In addition, we further evaluated MPXV-PCR-positive specimens by viral culture, electron microscopy, and viral inactivation assays. The limit of detection was established at 7.2 genome copies/reaction, and MPXV was successfully identified in 20 clinical specimens with 100% correlation against the reference method with 100% sensitivity and specificity. Our results demonstrated the validity of this rapid, robust, and reliable RT-PCR assay for specific and accurate diagnosis of MPXV infection in human specimens collected both as dry swabs and in viral transport media. This assay has been approved by NYS Department of Health for clinical use.
1 INTRODUCTION
Monkeypox virus (MPXV) is a zoonotic orthopoxvirus (OPXV) first isolated from skin lesions of cynomolgus macaques imported to Denmark in 1958.1, 2 This OPXV is endemic to areas of Central and West Africa, with occasional, but increasing numbers of reports outside the African continent.1, 3, 4 MPXV is typically transmitted to humans following exposure to a zoonotic source, with many animal species known to sustain the virus in nature.3 Although the main animal reservoir for MPXV remains unknown, many species of terrestrial/arboreal rodents,3, 5 nonhuman primates,3, 6 porcupines, and pigs have been reported to be naturally infected, contributing to its maintenance, potential zoonotic amplification, and repeated cross-overs into human populations.3, 7
The rapid geographical expansion of the virus and wide range of reservoir species seems to have broadened the human-MPXV interface.3 Additionally, cessation of smallpox vaccination, virus adaptation to new hosts in new regions, and increased contact between humans and MPXV-infected animals are potential factors contributing to the steadily increasing incidence of human-MPXV infections.3, 4 Recently, extended MPXV human-to-human transmission has been recognized in endemic areas of the Congo-basin with presumed enhanced transmission efficiency.8-10 Today, the ongoing intercontinental, multicountry outbreak supports these observations, suggesting the occurrence of cryptic and sustained virus transmission, presumably after the 2018−2019 outbreak followed by possible recent super-spreading events contributing further to abrupt worldwide dissemination.11, 12 The observation that the current MPXV-lineage (B.1) is a divergent descendent-branch from lineage A.1, responsible for the 2018−2019 outbreaks,11, 12 argues in favor of this scenario. Moreover, sustained human-to-human transmission may be shaping adaptation of currently circulating MPXV-strains, favoring evolution and conferring fitness advantages within human hosts, leading to increased transmissibility and infectiousness.
Transmission of MPXV occurs through close contact with cutaneous lesions,13 respiratory secretions,1, 14 via contact with contaminated surfaces/fomites,1, 15, 16 and reportedly through vertical transmission.17, 18 Clinical features of MPXV infection range from mild, nonspecific prodromal symptoms including fever, malaise, lymphadenopathy, and centrifugal macular and papulo-pustular rash, to severe systemic disease with respiratory or central nervous system manifestations.3, 13, 19-21 While infections with MPXV clades IIa (formerly known as West-African clade) tend to follow a milder course of disease, infections due to MPXV clade I (previously Central-African or Congo-Basin clade) are known to be more aggressive with higher case fatality rates, reported to be up to 10%.1, 3, 10, 22 Such differences in the clinical expression of disease are suggestive of distinct virulence factors occurring among strains, despite their narrow, ~0.5% genomic divergence.9, 23
MPXV infections may be difficult to diagnose clinically due to symptoms and signs that overlap both with noninfectious conditions and other cutaneous infections such as smallpox, chickenpox, rickettsialpox, disseminated herpes-zoster/herpes-simplex, eczema-herpeticum, scabies, measles, non-polio enteroviruses, syphilis, and endemic treponematosis.24-26 At the beginning of the current multicountry outbreak, case identification was hampered initially by their occurrence across a broad geography, often without a known link to endemic regions.13, 26, 27 Furthermore, continued spread has been maintained across communities via predominant occurrences in distinct social networks including men who have sex with men.28, 29
In light of the current rapid global spread of MPXV, there is an increasingly pressing need for highly specific diagnostic assays to facilitate identification of new cases, and to implement timely measures aimed at containing and preventing further transmission events.12, 30 Present guidelines from the World Health Organization (WHO) recommend using nucleic acid amplification tests (NAAT) for diagnosis, which can broadly detect OPXV or are specific for MPXV, with the latter being preferred.31 Currently, the United States Laboratory Response Network (LRN) has implemented an MPXV testing algorithm that utilizes the non-variola orthopoxvirus (NVO) real-time PCR (RT-PCR) that, if positive, requires further testing since it cannot differentiate MPXV from other OPXV.30
To circumvent this limitation, we evaluated and validated the performance of a multiplex RT-PCR assay (PKamp™ Monkeypox Virus RT-PCR RUO Kit; Perkin Elmer) to specifically and rapidly identify MPXV either from specimens collected in swabs transported dry or placed in universal/viral transport media (VTM). We report on the analytical sensitivity and specificity of this assay and demonstrate its utility as a swift and scalable diagnostic platform for identification of MPXV infection.
2 MATERIALS AND METHODS
This validation followed the guidelines and submission template for the rapid validation and approval of molecular assays for OPXV and/or MPXV diagnosis published by the New York Department of Health on July 1, 2022. The validation included testing the specificity, sensitivity, and analytic performance of the PKamp™ Monkeypox Virus RT-PCR RUO Kit32 in comparison to diagnostic results for dry swabs collected in parallel from the same lesions that were tested using the Centers for Disease Control (CDC) LRN Food & Drug Administration (FDA)-cleared NVO RT-PCR30 using the ABI 7500 Fast Dx Instrument performed by the New York City Public Health Laboratory at the Department of Health and Mental Hygiene (NYCPHL). A total of 40 specimens, each collected from a different patient, underwent testing by the reference method. These included 20 specimens that resulted as NVO-positive and 20 that resulted as NVO-negative by the reference method, which were evaluated on the laboratory-developed assay.
2.1 Target specificity
In the absence of reliable reference specimens from other OPXV, the analytical specificity of the assay was initially determined using an in silico approach for the primers-probe set included in the laboratory-developed assay.32 The primers/probes sequences are shown in Supporting Information: Table S1.
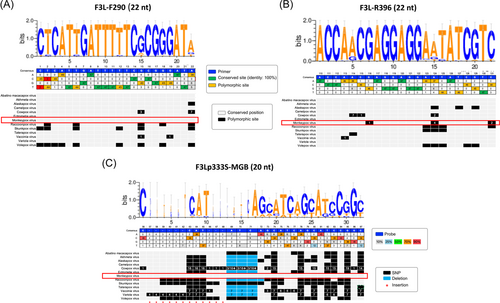
A total of 751 OPXV genomes were downloaded from the Global Initiative on Sharing Avian Influenza Data (GISAID, https://gisaid.org) and PubMed publicly-available databases for analysis. We performed a multiple sequence alignment with primers/probe sequences that target the viral F3L-gene (Table 1). The genomes analyzed comprised 13 viral species, which represented 47 distinct haplotypes at the primers/probe binding sites. From the haplotype alignment, the polymorphic sites for the primer/probe sequence regions were analyzed to evaluate how these variable sites could affect assay positivity.
| Viral species | Number of genomes | Number of haplotypes | F3L-F290 22 nt | F3L-R396 22 nt |
|---|---|---|---|---|
| Abatino macacapox virus | 2 | 1 | 22 (100%) | 22 (100%) |
| Akhmeta virus | 7 | 1 | 22 (100%) | 22 (100%) |
| Alaskapox virus | 1 | 1 | 21 (95.5%) | 19 (86.4%) |
| Camelpox virus | 10 | 2 | 22 (100%) | 22 (100%) |
| Cowpox virus | 91 | 19 | 20 (90.9%) | 19 (86.4%) |
| Ectromelia virus | 13 | 1 | 22 (100%) | 22 (100%) |
| Monkeypox virus | 394 | 4 | 22 (100%) | 19 (86.4%) |
| Raccoonpox virus | 3 | 1 | 17 (77.3%) | 18 (81.8% |
| Skunkpox virus | 2 | 1 | 16 (72.7%) | 19 (86.4%) |
| Taterapox virus | 2 | 1 | 22 (100%) | 21 (95.5%) |
| Vaccinia virus | 166 | 7 | 20 (90.9%) | 21 (95.5%) |
| Variola virus | 58 | 7 | 22 (100%) | 22 (100%) |
| Volepox virus | 2 | 1 | 13 (59.1%) | 17 (77.3%) |
| Total | 751 | 74 |
To evaluate the specificity of primer/probe sequences from the laboratory-developed assay against other known viruses that result in cutaneous exanthematic infections, we performed an in silico analysis on publicly-available viral genome sequences. Briefly, we utilized the NCBI blastn suite (https://blast.ncbi.nlm.nih.gov/) to align primer/probe sequences to viral genomes (partial and complete) in the NCBI Nucleotide collection, which is comprised of GenBank, EMBL, DDBJ, PDB, and RefSeq sequences. In addition, we aligned primer/probe sequences to six distinct taxa including Monkeypox (taxid: 10244), Herpes Simplex Virus Type 1 (HSV-1; taxid: 10298), Herpes Simplex Virus Type 2 (HSV-2; taxid: 10310), Varicella Zoster Virus (taxid: 10335), Enterovirus (taxid: 12059), and Human Parvovirus B19 (taxid: 10798) for a maximum of 500 target sequences using otherwise default criteria. We calculated average (and standard deviation) of the BLAST expectation value (e.g., E-value) across all alignments to infer the level of identity of each primer/probe sequences to 497 MPXV genomes (Supporting Information: Table S2).
To further evaluate the reliability of the assay, a specificity panel containing viral, bacterial, and fungal DNA from both clinical (viral) and culture (bacterial and fungal) samples was performed. The panel included: Herpes simplex virus Type 1, Herpes simplex type 2, Varicella zoster virus, Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus viridans, Serratia marcescens, Pseudomonas aeruginosa, Moraxella catarrhalis, Haemophilus influenza, Escherichia coli, Bacillus cereus, Nocardia farcinica, Mycobacterium chelonae, Candida glabrata, Candida guilliermondii, Candida kefyr, Candida krusei, Candida parapsilosis, Candida tropicalis, Penicillium spp., Fusarium spp., and Aspergillus niger.
2.2 Preparation, DNA extraction, and PCR assay
Dry swabs were resuspended in 1 ml of 50% ChemagicTM lysis buffer and vortexed for 30 s, whereas VTM collected-swabs underwent gentle shaking (200 rpm, 30 min, room temperature). A 300 µl aliquot was transferred from each specimen, to individual wells of a 2 ml deep-well-plate, then 300 μl of lysis buffer and extraction master mix (4 μl Poly(A) + 10 μl Proteinase K) was added to each well.
DNA was extracted using the ChemagicTM Viral DNA/RNA 300 Kit H96 (CMG-1033-S; Perkin Elmer) on the automated ChemagicTM 360 instrument (2024-0020; Perkin Elmer) as per manufacturer's protocol and as previously described.32 The laboratory-developed RT-PCR assay was conducted in a Real-Time PCR system Bio-Rad CFX 384-Well Thermal Cycler (Bio-Rad Laboratories) using the PKamp™ MPXV detection kit V1 (Perkin Elmer). The reaction mixture contained 3.75 μl of Reagent A (Buffers, dNTPs, Mg2+), 0.75 μl of MPXV Reagent B1 (TE buffer, primer pair, and probe), 0.5 μl of enzyme mix (Taq DNA polymerase, UNG), and 10 μl of extracted nucleic acid. Thermal cycling conditions were as follows: 2 min at 37°C (1 cycle), 10 min at 94°C (1 cycle), and 40 cycles of 10 s (94°C) and 1 min (63°C).
2.3 Analytic sensitivity
The limit of detection (LoD) of the laboratory-developed assay was determined using the positive control from the PKamp™ Monkeypox Virus RT-PCR RUO Kit. This positive control is a plasmid construct containing the Monkeypox-specific F3L gene and the human RNase P gene (as endogenous control) with a stock concentration of 20 000 copies/reaction. Ten-fold serial dilutions were generated by spiking pooled clinical matrices (e.g., dry swabs or in VTM) from specimens that tested negative for MPXV by the reference method. Three replicates at each dilution were run by three independent technologists on three different runs (e.g., days).
Linear regression analyses were performed across the cycle-threshold (Ct) values resulted for each replicate at each serial dilution. Intra- and inter-assay differences in Ct-values across the replicates tested were statistically evaluated using R-software. For continuous values, Shapiro−Wilk test was used to test normality. The nonparametric Kruskal−Wallis test was used to compare nonparametric distributions. All tests were two-tailed, and p < 0.05 considered statistically significant. To statistically evaluate the LoD, a Probit regression was conducted.
2.4 Reproducibility
Inter- and intra-assay reproducibility across clinical specimens was assessed using DNA from six specimens that resulted as NVO-positive (n = 3) or NVO-negative (n = 3) by the reference method. Each was run in triplicate by three independent technologists on three different runs (e.g., days). To evaluate if there were statistically significant intra- or inter-run differences, statistical analyses were carried out using R-software. Statistical tests were applied as described above.
2.5 Diagnostic performance
To evaluate the diagnostic performance of the PKamp™Monkeypox Virus RT-PCR RUO Kit, we tested 20 NVO-negative and 20 NVO-positive specimens (Table 2) resulted by the reference method. We calculated the diagnostic sensitivity and specificity, negative predictive value (NPV), positive predictive value (PPV), and level of interrater agreement (e.g., kappa index).
| Sample ID | Age (years) | Gender | Site | Non-variola OPXV DNAa | MPXV Ctb | RNase P Ctc | Sample typed | Culture resulte |
|---|---|---|---|---|---|---|---|---|
| 1 | 28 | Male | Anorectal | Detected | 18.82 | 25.67 | Dry swab | Growth |
| 19.03 | 26.58 | VTM | ||||||
| 2 | 46 | Male | Lesion unspecified | Detected | 16.43 | 27.97 | Dry swab | Growth |
| 18.68 | 30.05 | VTM | ||||||
| 3 | 38 | Male | Penile lesion | Detected | 24.54 | 28.85 | Dry swab | Growth |
| 19.02 | 28.35 | VTM | ||||||
| 4 | 40 | Male | Lesion unspecified | Detected | 24.18 | 30.11 | Dry swab | Growth |
| 24.97 | 32.18 | VTM | ||||||
| 5 | 36 | Male | Lesion unspecified | Detected | 18.59 | 25.32 | Dry swab | Growth |
| 17.89 | 26.07 | VTM | ||||||
| 6 | 44 | Male | Left temple | Detected | 18.29 | 24.59 | Dry swab | Growth |
| 20.23 | 27.80 | VTM | ||||||
| 7 | 28 | Male | Right third digit | Detected | 31.80 | 35.19 | Dry swab | No growth |
| 34.01 | 36.50 | VTM | ||||||
| 8 | 34 | Male | Anorectal | Detected | 16.55 | 23.41 | Dry swab | Not tested |
| 16.64 | 22.27 | VTM | ||||||
| 9 | 35 | Male | Lesion unspecified | Detected | 17.68 | 27.62 | Dry swab | Growth |
| 17.38 | 29.07 | VTM | ||||||
| 10 | 34 | Male | Lesion unspecified | Detected | 22.42 | 27.42 | Dry swab | Growth |
| 18.57 | 25.51 | VTM | ||||||
| 11 | 27 | Male | Right chest | Detected | 18.87 | 26.18 | Dry swab | Growth |
| 17.85 | 25.62 | VTM | ||||||
| 12 | 25 | Male | Anterior penile lesion | Detected | 20.24 | 26.81 | Dry swab | Growth |
| 20.28 | 26.95 | VTM | ||||||
| 13 | 31 | Male | Anorectal | Detected | 19.68 | 30.73 | Dry swab | Growth |
| 21.57 | 31.20 | VTM | ||||||
| 14 | 54 | Male | Right shoulder | Detected | 17.42 | 22.99 | Dry swab | Growth |
| 20.09 | 25.34 | VTM | ||||||
| 15 | 46 | Male | Left hand | Detected | 15.71 | 22.80 | Dry swab | Growth |
| 17.72 | 24.45 | VTM | ||||||
| 16 | 48 | Male | Scrotal | Detected | 28.54 | 32.22 | Dry swab | Growth |
| 27.40 | 32.58 | VTM | ||||||
| 17 | 37 | Male | Perioral | Detected | 20.45 | 27.25 | Dry swab | Growth |
| 20.26 | 31.35 | VTM | ||||||
| 18 | 30 | Male | Anorectal | Detected | 25.16 | 29.94 | Dry swab | Growth |
| 22.63 | 30.18 | VTM | ||||||
| 19 | 34 | Male | Penile lesion | Detected | 21.66 | 26.58 | Dry swab | Growth |
| 21.52 | 26.69 | VTM | ||||||
| 20 | 49 | Male | Penile lesion | Detected | 14.13 | 23.26 | Dry swab | Growth |
| 11.34 | 20.81 | VTM |
- a Result from non-variola orthopoxvirus (OPXV) real-time PCR (NYCPHL).
- b Cycle threshold (Ct) of monkeypox virus (MPXV) DNA by laboratory-developed assay.
- c Cycle threshold (Ct) of endogenous control DNA, RNase P, by laboratory-developed assay.
- d Side-by-side specimens collected via a dry swab versus a swab of the same cutaneous lesions in viral transport media (VTM).
- e In vitro culture result of human or monkey cell lines inoculated with VTM-collected specimen.
2.6 In vitro cell culture, virus inactivation, and electron microscopy
All work with viral cultures was performed in a the BSL-3 Conventional Biocontainment facility at ISMMS by trained personnel using standard operating procedures approved by the Mount Sinai Institutional Biosafety Committee.
MPXV underwent in vitro cell culture by infecting dermal fibroblasts immortalized by hTert transduction33 or BSC-40 cells. Infected cells were assessed for cytopathic effect (CPE). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCSi, 56°C, 30 min). BSC-40 cells (African green monkey kidney cells) were cultured in DMEM 5% FCSi. Cells were cultured at 37°C and 10% CO2, and tested for Mycoplasma contamination using the PlasmoTest kit according to the manufacturer's instructions (Invivogen).
Monolayers of BSC-40 or dermal fibroblasts (plated in 12-well plates) were infected with 100 μl of viral transport media. After 1 h incubation at 37°C and 5% CO2, cells were washed and incubated with DMEM supplemented with 2% FCS, for 24 or 48 h and CPE and plaques were visualized using EVOS M5000 Imaging System (Thermo Fisher Scientific).
To assess whether the lysis buffer used for the DNA extraction fully inactivates the virus, a clinical specimen (PV67624) with low Ct values (MPX Ct: 15.8) was mixed with ChemagicTM DNA/RNA lysis buffer or VTM and used to infect dermal fibroblasts as described above. Lysis buffer alone was included in addition to mock controls. Cells were inspected for signs of CPE at 48 h postinfection (hpi).
Monolayers of human fibroblasts were infected in six-well plates with 100 μl of PV67624 as described above in the BSL-3 facility. CPE was evident at 9 hpi, and supernatant was removed. Harvested cells underwent primary fixation in 3% codylate buffered glutaraldehyde and secondary fixation in 1% codylate buffered osmium tetroxide. Pelleted specimens were dehydrated in graded ethanol and embedded in epoxy media in standard fashion. One micron methylene-blue stained scout sections were prepared, followed by 90 nm ultrathin sections. Ultrathin sections were stained with Uranyless proprietary stain (Electron Microscopy Sciences) and lead citrate. Sections were examined in a Hitachi H7800 TEM at 80 kV.
3 RESULTS
We evaluated the analytical specificity of the laboratory-developed assay through an in silico approach. Table 1 summarizes the information of the genome sequences included, the number of haplotypes, and the identity percentages across primer/probe sequence regions for each OPXV evaluated. Figure 1 illustrates the predominant nucleotide by position in the alignment (upper section of each panel) and which OPXV demonstrate polymorphisms by position (Lower section of each panel). We noted that the annealing regions of the primers were highly conserved amongst different OPXV, with the probe revealing the highest specificity. Additional analyses contrasting MPXV against other viruses including HSV-1, HSV-2, VZV, Enterovirus, and Human Parvovirus B19 confirmed a high level of specificity for these primers/probe sequences to MPXV genome sequences (Supporting Information: Table S2). In addition, the PCR testing using a panel containing viral, bacterial, and fungal DNA from both clinical (viral) and culture (bacterial and fungal) samples showed negative results (Supporting Information: Table S3). Lastly, due to the recent CDC alert “Lab Alert: MPXV TNF Receptor Gene Deletion May Lead to False Negative Results with Some MPXV Specific LDTs,” we decided to download the available MPXV genomes from the B.1 lineage and evaluated if there were any mismatches in the annealing sites of the primers and the probes. Our results did not detect any mismatches suggesting the reliability of the primers/probe set for MPXV detection by this assay (Supporting Information: Figure S1).
We next evaluated the analytic performance of the laboratory-developed assay. Figure 2A,B show the linear regression and dynamic range of the LoD experiments. We found the lowest concentration at which we detect MPXV in 100% of (e.g., 3 of 3) replicates was 20 copies/reaction. The LoD was calculated as the lowest dilution providing 95% positive results, as established by Clinical Laboratory Standards Institute standards by means of the Probit regression model. An LoD of 7.20 MPXV genome copies/reaction (95% confidence interval = 5.11−9.30) was determined for both VTM and dry swab specimens (Figure 2C,D).
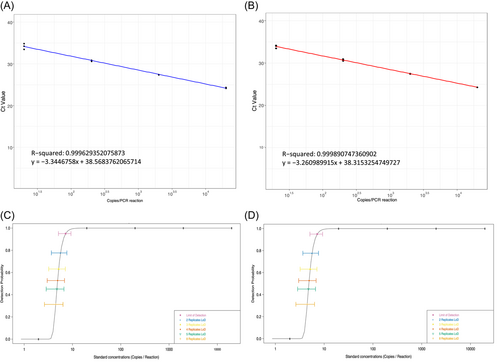
The Ct-values of the triplicate replicates run across three independent runs are shown on Supporting Information: Table S3. There were no statistically significant differences in Ct-values across runs and replicates or across specimen matrix types (χ2 = 0.73135, df = 2, p = 0.6937; χ2 = 0.24, df = 2, p = 0.8869).
We also performed intra- and inter-run experiments using clinical specimens and found no statistically significant intra- or inter-run differences which further demonstrates a high level of reproducibility (χ2 = 0.42, df = 2, p = 0.8106). Finally, among the 40 clinical specimens tested on the laboratory-developed assay, we found all 20 specimens that yielded NVO-positive results by the reference method were MPXV-positive on the laboratory-developed assay. Moreover, all 20 specimens that yielded NVO-negative results by the reference method were MPXV-negative on the laboratory-developed assay. Together, this demonstrates a 100% diagnostic sensitivity, diagnostic specificity, NPV, PPV, and kappa index. Furthermore, this was consistent across both specimens collected in VTM or as dry swabs.
Importantly, we performed in vitro viral cultures using VTM from 19 of the 20 positive swabs to evaluate the infectivity of each clinical specimen in comparison with laboratory-developed assay results. Inoculation of cells with 18/19 clinical specimens resulted in characteristic cytopathic changes at 2 days postinfection. Specimen PV67607 did not yield infectious virus, which is in good agreement with the fact that it had the lowest amount of detectable MPXV (Ct: 34.01). A summary of laboratory analysis and virological screening including MPXV RT-PCR versus Orthopoxvirus generic RT-PCR assays and cultures are shown in Table 2.
We next experimentally determine whether the lysis buffer used for the DNA extraction efficiently inactivates MPXV. VTM from PV67624 not treated with lysis buffer developed CPE at 24 hpi (Figure 3A,B) while the same sample mixed with lysis buffer failed to produce CPE in cell culture after 24−48 hpi while retaining cell viability (Figure 3J−L).
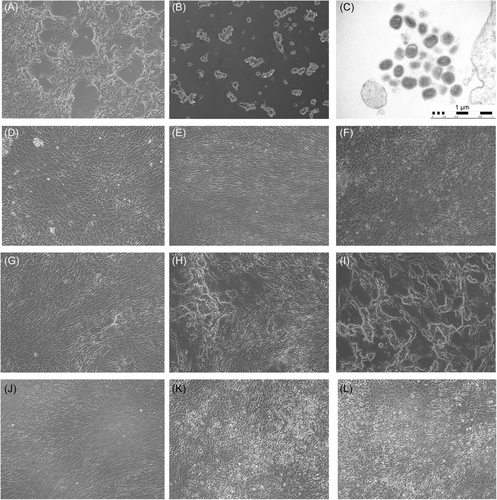
Electron microscopy assessment generated using infected cells showed distinct poxvirus ultrastructural findings indicating the presence of different stages of viral morphogenesis (Figure 3C).
4 DISCUSSION
Since 2017, monkeypox cases have re-emerged causing outbreaks across endemic regions of Africa, particularly Nigeria, which has raised concerns about the potential spread of MPXV to other regions and hosts beyond the African borders.3, 4, 26 Indeed, since May 2022, the world has witnessed an unprecedented rise of MPXV cases affecting 89 WHO member-states (https://worldhealthorg.shinyapps.io/mpx_global/, as of August 9, 2022) across different geographical areas, and with no clear epidemiological link to endemic regions nor apparent evidence of zoonotic transmission.34 This ongoing global outbreak has reached >30 000 laboratory-confirmed cases worldwide following sustained human-to-human transmission, thus signaling the potential occurrence of adaptive changes in MPXV that may lead to enhanced infectivity and transmission.11, 12 In response, the WHO has declared the current outbreak a global health emergency,34 and most public health authorities across the world have implemented early screening for suspicious cases by ramping up testing efforts.30, 35 Indeed, reference centers such as the US CDC and other public health and commercial laboratories have partnered under the LRN to collectively ensure adequate laboratory infrastructure and preparedness.30
Given this, access to diagnostic tests remains critical to capture new infections to, in turn, inform early mitigation measures aimed at rapidly and effectively preventing further viral spread. A number of diagnostic methods and assays have been described for the detection of MPXV, including viral culture, antigen detection, and serological tests with variable performance.36 However, over the last decade, molecular diagnostics including NAAT and DNA sequencing assays have been developed as these technologies have become more readily available and utilized.37-40 The advantage of these molecular approaches is that they allow detecting viral gene targets from a variety of biological samples, including both primary (e.g., cutaneous lesions swabs, blood, and semen) and environmental specimens.15, 16, 19, 41
Fully automated sample-to-result systems are under development for MPXV testing. One example is the GeneXpert MPX/OPX platform which combines an MPXV-specific and OPXV-generic multiplex approach with 98.8% sensitivity and 100% specificity, reported both under laboratory and field conditions.42 Such platforms have the advantage for scalability and high-throughput testing efforts. However, presently the use of the GeneXpert system may be cost-prohibitive compared to conventional RT-PCR platforms when used for routine diagnosis.
RT-PCR is the most common approach to detect MPXV in the laboratory and is considered the diagnostic gold standard.30 These assays usually vary in the viral genes targeted, sensitivity, specificity, and level of detection. The PKamp™ Monkeypox Virus RT-PCR from Perkin Elmer, available for research use only (RUO), uses species-specific oligonucleotide primers and probes targeting the F3L-region of the MPXV genome (a.k.a OPG065),43, 44 offering a rapid an accurate method to undisputedly diagnose MPXV within the broader range of other OPXV.
Here, through in silico approaches, we demonstrate that this assay has a high specificity to MPXV among a broad set of diverse OPXV. Importantly, we show the assay's primer/probe sequences have high specificity to historical and contemporary MPXV-genomes and minimal complementarity to pathogens that cause common vesiculopustular exanthematic lesions including HSV-1, HSV-2, VZV, enterovirus (e.g., Coxsackievirus), and parvovirus B19 (Figure 1; Table 1). In addition, the assay tested negative when using a panel of viral, bacterial, and fungal pathogens that might present differential diagnosis with MPXV (Supporting Information: Table S3). Despite the recent CDC alert “Lab Alert: MPXV TNF Receptor Gene Deletion May Lead to False Negative Results with Some MPXV Specific LDTs,” the primers and probe used in this assay do not have annealing mismatches so far, that might result in potential false negatives. Nevertheless, continuing genomic surveillance is needed to foresee potential mutations affecting MPXV NAAT assays worldwide (Supporting Information: Figure S1).
In our validation experiments, this assay displayed a high level of accuracy in comparison to an established, FDA-cleared reference method. Indeed, there was perfect concordance between a set of NVO-positive and -negative specimens run on the assay presented (Table 2). Moreover, this robust performance was equally reflected across two distinct specimen matrices: dry swabs and VTM. Together, these qualities are essential to accurately diagnose new MPXV infections particularly as MXPV continues to spread among wider communities, and thus, needs to be identified from other, clinical dermatologic presentations.
We also demonstrate that this assay has a high level of analytic sensitivity with an estimated LoD at 7.20 genome copies/reaction and is reliably reproducible (Figure 2). Indeed, this assay was able to detect MPXV across a variety of anatomic sites that span the manifestation of early-to-late stages of MPXV infection. Furthermore, these clinical specimens yielded a range of viral quantities as seen by a range of Ct-values from 14.13 to 31.80 for dry swab specimens and 11.34−34.01 for VTM swab specimens. As we attempted to correlate growth in viral culture with Ct-values as markers for quantity inoculated, we can preliminarily infer that a Ct >30 may be associated with the cut-off for in vitro infectivity. However, more studies are warranted to better understand viral dynamics and establish reliable Ct threshold to inform disease status and infectious risk.
Confirmation by electron microscopy (Figure 3), the traditional gold standard for identifying novel and emerging viruses, further refined diagnosis at an ultrastructural level by revealing characteristic poxvirus virions with their brick-shaped appearance, widely defined envelope with electron-dense core and whorled surface.
Given the importance and safety implications of adequate viral inactivation, we additionally evaluated the ability of the lysis buffer to halt viral infectiousness by inoculating onto cell cultures virus-buffer mixtures from a selected sample. The complete absence of visible CPE and lack of structural changes that indicate cell toxicity after 24−48 h of incubation following inoculation with lysis buffer-treated specimens confirmed the capability of the lysis buffer to inactivate viral samples and thus ensures safe handling and processing of the samples by laboratory personnel.
An additional advantage of this assay relies on the fact that it lends itself to be easily adapted to automated-systems such as the Perkin Elmer automated nucleic acid solutions platform, allowing optimal extraction and minimal handling by using automated liquid handling workstations. In all, this provides a higher quantity and scalable throughput and rapid turnaround time much needed to inform infection control strategies in context of outbreak conditions such as the current MPXV epidemic. Furthermore, minimal sample manipulation reduces the risks for exposure and contamination for laboratory staff.
Overall, we found the PKamp™ Monkeypox Virus RT-PCR RUO-assay to be a reliable, specific, and sensitive test for detecting MPXV infection in humans. This assay has been approved by the NYS Department of Health for clinical use as Laboratory Developed Test, and offers an alternative for testing in context of the current multicountry outbreak were diagnostic assays are of utmost relevance worldwide.
AUTHOR CONTRIBUTIONS
Alberto Paniz-Mondolfi, Susana Guerra, and Juan David Ramírez: conceptualization, study design, methodology, formal analysis, writing original draft, and final review. Marina Muñoz, Nicolas Luna, and Jason Reidy: investigation, data curation, and methodology. Matthew M. Hernandez: data curation, methodology, writing review, and editing. Radhika Banu, Paras Shrestha, Bernadette Liggayu, and Audrey Umeaku: supervision, project administration, and resources. Feng Chen, Liyong Cao, Armi Patel, Ayman Hanna, Sunny Li, and Nina Pagani: validation, investigation, and data collection. Randy Albrecht, Rebecca Pearl, Adolfo Garcia-Sastre, Dusan Bogunovic, Gustavo Palacios, Harm van Bakel, Ana Gonzalez-Reiche, Viviana Simon, and Emilia Mia Sordillo: methodology, writing review, and editing. Lucia Bonnier, Freddy Cera, Heidi Lopez, Yvette Calderon, Erick Eiting, Karr Mullen, Sangyoon Jason Shin, Luz Amarilis Lugo, Antonio E. Urbina, Carlotta Starks, Tonny Koo, Patricia Uychiat, and Avery Look: investigation, data collection, and supervision. Adolfo Firpo Betancourt, David Reich, and Carlos Cordon-Cardo: project administration, funding acquisition, writing review, and editing.
CONFLICTS OF INTEREST
The A. G.-S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapuetics, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories and Merck, outside of the reported work. A. G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, Paratus, CureLab Oncology, CureLab Veterinary, Synairgen and Pfizer, outside of the reported work. A. G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott and Astrazeneca. A. G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. D. B. is a founder and part owner of Lab11 Therapeutics Inc. The remaining authors declare no conflict of interest.
ETHICS STATEMENT
Residual clinical specimens were utilized for this study as part of the Mount Sinai Pathogen Surveillance Program, which has been reviewed and approved by the Human Research Protection Program at the Icahn School of Medicine at Mount Sinai (ISMMS) (HS#13-00981).
Open Research
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.




