Evidence for human infection with avian influenza A(H9N2) virus via environmental transmission inside live poultry market in Xiamen, China
Yifei Jin, Huan Cui, and Lina Jiang contributed equally to this study.
Abstract
H9N2 avian influenza virus (AIV) has become prevalent in the live poultry market (LPM) worldwide, and environmental transmission mode is an important way for AIVs to infect human beings in the LPM. To find evidence of human infection with the influenza A(H9N2) virus via environmental contamination, we evaluated one human isolate and three environmental isolates inside LPMs in Xiamen, China. The phylogeny, transmissibility, and pathogenicity of the four isolates were sorted out systematically. As for the H9N2 virus, which evolved alongside the “Avian-Environment-Human” spreading chain in LPMs from the summer of 2019 to the summer of 2020, its overall efficiency of contact and aerosol transmissibility improved, which might contribute to the increasing probability of human infection. This study indicated that environmental exposure might act as an important source of human infection in LPMs.
1 INTRODUCTION
Since the first human infection with avian influenza A(H9N2) virus was identified in 1999,1 a total of 110 human cases have been detected worldwide. As of March 30, 2022, China has experienced 97 epidemics of human infections with the avian influenza A(H9N2) virus, which are mainly concentrated in southern China.2 Risk factors for human infection with H9N2 virus mainly include exposure to the live poultry market (LPM) and raising backyard poultry, so poultry workers are one of the most likely infected groups.
According to the Influenza Risk Assessment Tool (IRAT) assessment results, the potential pandemic risk of H9N2 is only less than that of H7N9 highly pathogenic avian influenza virus (HPAIV) and H5N6 HPAIV.3 H9N2 virus has gradually become the dominant strain in the “Avian-Environment-Human” source in LPMs.4 Concern exists that the presence of H9N2-carrying poultry might act as an incubator that could introduce new influenza subtypes into the human population through a process of genetic reassortment and mutation, or both.5 It has been observed that H9N2 is more infectious to humans than other AIVs.6 In China, the H9N2 infection risks of poultry workers in LPM were higher than those of the general population.7
The environmental transmission mode is an important way for AIVs to infect human beings in LPM.8-11 The LPM, which is composed of different pathogenic resources, can easily lead to the circular transmission of AIVs.12 Thus, it is necessary to perform continuous monitoring for the evolution of the H9N2 virus during the process of environmental transmission. In this study, to find evidence for human infection with avian influenza A(H9N2) virus via environmental contamination, we evaluated one isolate from a patient and three from the environment inside LPMs in terms of genetic evolution and virological characteristics containing transmissibility and pathogenicity. As they evolved, their efficiency of contact and aerosol transmissibility increased. Those data indicated that environmental exposure might become the source of human infection.
2 METHODS
2.1 Sample collection and virus isolation
Patients with respiratory tract specimens that were positive for H9 and N2 based on real-time reverse transcription polymerase chain reaction were confirmed as H9N2 infections through the Xiamen City Center for Disease Control and Prevention of China. We collected one positive throat swab from a patient and three positive environmental samples from two relevant LPMs in the same district. H9N2 viruses were isolated in biosafety laboratories by using 10-day specific pathogen-free (SPF) embryonated chicken eggs. The hemagglutinin (HA) activity was assayed for the allantoic fluid.
2.2 Genome sequencing and phylogenetic analysis
Viral RNAs were extracted from HA-positive allantoic fluid by using QIAamp Viral RNA Mini Kit (QIAGEN). Viral RNAs were reverse-transcribed into cDNA by using the primer Uni12 and RT reagent Kit (Takara). The obtained cDNA was used for specific amplification and viral genome sequencing (Shanghai Sangon Biotech Co., Ltd.).
All reference sequences used in this study were obtained from the National Center for Biotechnology Information GenBank database, and then the downloaded sequences were compared with the strains in this study through Cluster W. The GTRGAMMA nucleotide substitution model in PhyML 3.1 software was used and bootstrap replicates were run 1000 to evaluate the maximum likelihood (ML) phylogenies of codon comparison between the two gene sequences. The phylogenetic relationship of the HA gene of four strains was inferred with BEAST v1.10.4 by using a molecular clock that placed a timescale on virus evolution to estimate rates of viral evolution and dates of divergence.13 Phylogeny is estimated within the framework of the Bayesian Markov chain Monte Carlo (MCMC).14 A maximum clade credibility (MCC) tree with mean height was generated for each data set by using TreeAnnotator version 1.10.4 (https://beast.community/treeannotator) after 10% burn-in. In addition, at least two independent runs were carried out and compared to ensure sufficient sampling. The phylogenetic tree and MCC tree were visualized by using FigTree version 1.4.4 (http://tree.bio.ed.ac.uk).
2.3 Guinea pig experiments
Hartley strain female guinea pigs weighing 300–350 g (Vital River Laboratory Animal Technology Co., Ltd.) were used in this study. Different infection groups (three guinea pigs) were inoculated intranasally with 106 EID50 of the required strains in a 200 µl volume (100 µl per nostril), and then housed in a cage inside an isolator. Three naïve guinea pigs per group were cohoused (in the same cage) with the three infected guinea pigs on 1-day postinoculation to study direct-contact transmission, and another three naïve guinea pigs per group were housed in adjacent cages (5 cm apart), to study aerosol transmission. Nasal wash samples were collected on 1-, 3-, 5- and 7-days postinoculation. Serum was collected from all guinea pigs on 21-days postinoculation and the HI test was performed according to the protocol described in the World Health Organization guidelines.15 The human-isolate A/California/04/2009 (CA04[H1N1]) was used as the reference strain for this study.
2.4 Growth dynamics in cells
The growth kinetics of four strains in A549 and MDCK cells were compared according to the protocol of the previous study.16 Each strain was inoculated into monolayers of A549 or MDCK cells at a multiplicity of infection (MOI) of 0.001. Then we collected the supernatant of each well at a different time. Supernatants of infected cells were collected at 12, 24, 36, 48, and 60 h postinfection and stored at −80°C, and the virus titer of each collected sample was determined by EID50 assays.
2.5 Mouse experiments
The pathogenicity of each H9N2 strain in mammals was evaluated by mouse experiments. Six-week-old BALB/c female mice (Vital River Laboratory Animal Technology Co., Ltd.) were used in this study. The mice infection experiments were divided into two parts. The first part is weight monitoring, and 25 mice were equally divided into five groups, including four infection groups and one control group, which were anesthetized with isoflurane and inoculated nasally with 50 µl different H9N2 strain at 106 EID50 and 50 µl PBS, respectively. All mice body weights were continuously monitored for 14 days. The second part is the monitoring of virus replication and pathogenicity in vivo. A total of 85 mice were randomly divided into five groups, four infection groups (20 mice) were infected with different H9N2 strains at 106 EID50, and one group (five mice) was mock-infected with PBS. Each infection group with five mice that were euthanized on 1-, 3-, 5-, and 7-days postinoculation. A total of 10 tissues (heart, liver, spleen, lung, kidney, brain, trachea, pancreas, intestine, and turbinate) were collected and separately homogenized in 1 ml of PBS. Viral growth in each tissue was determined by EID50 assays. H&E staining of mice lung were performed on 3-days postinoculation.
2.6 Statistical analysis
One-way analysis of variance (ANOVA) was performed using GraphPad Prism 8.0 to determine statistically significant differences. All analyses were performed in triplicate and are representative of at least three separate experiments. Error bars represent standard deviation.
3 RESULTS
3.1 Epidemiological investigation and phylogenetic analysis
On January 3, 2021, a 52-year-old female worker in Zhenhai LPM of Siming District in Xiamen, went to the hospital for treatment due to fever. The clinical test results of the patient on January 3, 2021: white blood cell counts 17.71 × 109/L, neutrophil ratio of 74.60%, and lymphocyte ratio of 17.30%. The results of chest computed tomography (CT) scan: (1) Bilateral pulmonary bronchiectasis with infection, pleural thickening, mediastinum and hilar lymph node enlargement, mediastinal structure moving to the right; (2) right upper lobe lesion; (3) tracheal diverticulum (Supporting Information: Figure S1). Her respiratory tract sample was tested positive for influenza A(H9N2) at Xiamen Center for Disease Control and Prevention. We collected 21 environmental samples in the two relevant LPMs, including eight samples from Zhenhai LPM and 13 samples from Punan LPM. Subsequently, three positive environmental samples were collected in two relevant LPMs. Viruses in the four samples were successfully isolated in embryonated chicken eggs (GenBank: ON856627-ON856634). A/Fujiansiming/19/2021 (abbreviated as H19) was isolated from the patient. A/Environment/Xiamen/01/2021 (abbreviated as E01) was isolated from the sample wiped on the surface of the cutting board of Zhenhai LPM. A/Environment/Xiamen/02/2021 (abbreviated as E02) and A/Environment/Xiamen/03/2021 (abbreviated as E03) were isolated from wiping samples on the surface of the cutting board and poultry drinking water (PDW) of Punan LPM in the same district, respectively (Figure 1A).
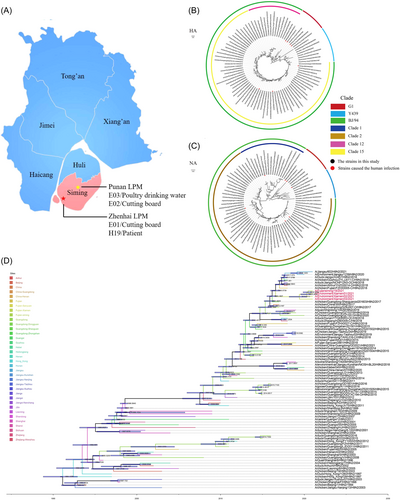
To understand their evolutionary relationship, we performed sequencing and phylogenetic analysis of viral genomes. We found that HA and NA genes of four strains were closely related to reference strain A/chicken/Anhui/YHZS014-O/H9N2/2018 and A/chicken/Fujian/FZHX0030-C/H9N2/2018, respectively (Figure 1B,C). To further characterize virus evolution, Bayesian time-resolved phylogenetic tree was mapped, showing that all four strains had an obvious time evolutionary relationship and were close to those in Guangzhou and Fujian, China (Figure 1D). In general, E03 was separated in the summer of 2019 and was the earliest of four strains in this study. E02 was separated from E03 in the spring of 2020. E01 and H19 were separated from E02 in the summer of 2020. Based on the evolutionary relationship among the four strains, we further explored their differences in transmissibility and pathogenicity.
3.2 Determination of transmissibility and pathogenicity of viruses
The receptor-binding specificity of each strain was determined by HA assays (Supporting information). Results showed that each of the four strains exhibited a binding affinity for desialylated cRBCs that expressed SAα-2,6-Gal, suggesting that all four strains had the risk of cross-species infection (Supporting Information: Figure S2). We then evaluated the contact and aerosol transmission efficiency of four strains in guinea pigs. For contact transmission efficiency, it decreased from 67% (E03 in summer 2019) to 33% (E02 in spring 2020), and then increased to 100% (E01 and H19 in summer 2020) (Figure 2A). For aerosol transmission efficiency, it gradually increased from 0% (E03 in summer 2019 and E02 in spring 2020) to 33% (E01 in summer 2020) and 100% (H19 in summer 2020) (Figure 2A). These data suggest that the overall transmission efficiency of those H9N2 viruses gradually increased in the process of evolutionary adaptation. Serological results corresponded to virus detection experiments (Supporting Information: Figures S3 and S4).
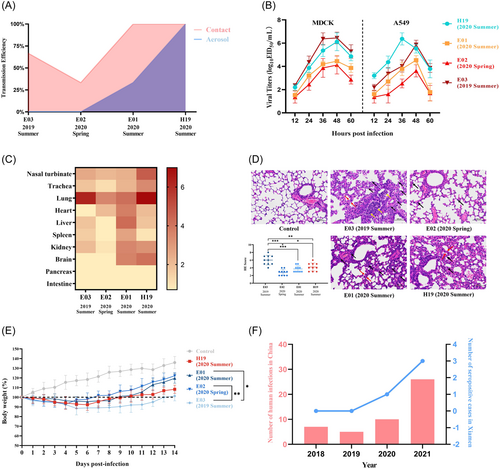
Since the transmissibility of the four strains had been enhanced with the evolution of the virus, we wondered whether the replication ability of the four strains also had a certain changing trend. The results showed that E03 and H19 had the strongest proliferation ability on MDCK cells and A549 cells, respectively (Figure 2B). We further evaluated the virulence by intranasal inoculation with 106 EID50 of the virus in BALB/c mice (Figure 2C). In addition to the high viral titer of E03 in the lung, the viral titers in other organs, including the upper respiratory tracts, were low. E02 was low in all organs while E01 increased widely among organs except the pancreas and intestine (Figure 2C). The viral titer of H19 in the upper and lower respiratory tract increased significantly (Figure 2C). Besides, E01 and H19 could be detected in the brain (Figure 2C and Supporting Information: Figure S5), indicating they acquired the ability to cross blood–brain barrier. These results showed that in upper respiratory tract (nasal turbinate and trachea) and lower respiratory tract (lung), the changing trend of virus titer is consistent with the transmission efficiency. It demonstrated that E01 might acquire transmissibility from the environment to humans. To determine pathological damage, pulmonary tissues were harvested from mice on 3-days postinoculation and stained with H&E. The degree of damage was highest in E03, followed by E01 and H19, and lowest in E02, which is consistent with the results of viral titers in lungs (Figure 2D). After 14 days of bodyweight measurement and death monitoring, we found that the infected mice had no noticeable clinical symptoms but weight loss (Figure 2E). The maximum weight change was observed in E03-infected mice, while the minimum weight change was observed in E02-infected mice. The weight changes of mice with E01 or H19 infection in 1–10 days were similar, but H19 was recovered slowly in 11–14 days. Based on the above results, we found that E01 was closest to H19 in terms of contact transmission and aerosol transmission efficiency, which also indicated that E01 had the potential to infect humans. However, E01 was different from H19 in terms of pathogenicity to mammals, which might be the result of virus adaptation to human.
4 DISCUSSION
The environmental transmission mode is an important way for AIVs to infect human beings in the LPMs, which has the potential to cause the circular transmission of influenza A viruses. The viability of the AIVs in the environment is determined by the combined effects of multiple chemical and physical factors, such as humidity, temperature, pH, salinity, and organic compounds, as well as its own properties.17-19 According to the statistics of the China Meteorological Data Service Centre, the typical annual average temperature in Xiamen is 21°C, and the annual average relative humidity is 78%.20 Therefore, H9N2 has certain survivability in this climate environment, which provides more opportunities for its evolution and mutation.
By investigating the number of human infection cases with H9N2 virus in China from 2018 to 20212 and the number of poultry workers that were seropositive for H9N2 virus from LPMs in Xiamen, we found that the number of patients with H9N2 infection increased from 2018 to 2021 (Figure 2F), which is consistent with the prior reports.4, 5 The number of H9N2-seropositive poultry workers in Xiamen also broke the record of no infection since 2020, when the transmissibility of E01 virus significantly increased. As the H9N2 virus evolved alongside the “Avian-Environment-Human” spreading chain in LPMs, its overall efficiency of contact and aerosol transmissibility improved, which might have contributed to the increasing probability of human infection. This study indicated that environmental exposure might act as a source of human infection. Our study still has one limitation because the monitoring sample size needs to be further improved.
Overall, we found evidence of human infection with the avian influenza A(H9N2) virus via environmental contamination inside LPM in Xiamen, China. It is suggested to strengthen environmental disinfection management to solve the circular transmission of AIVs in the LPM and to prevent cross-species infection and human pandemic.
AUTHOR CONTRIBUTIONS
Zhongyi Wang, Bing Lu, and Zhendong Guo initiated and coordinated the project. Yifei Jin, Huan Cui, and Lina Jiang designed the experiments. Yifei Jin, Lina Jiang, Zehui Chen, Jing Zheng, Yidun Zhang, and Li Li collected samples and isolated viruses. Huan Cui, Cheng Zhang, and Zhendong Guo conducted the cell and animal experiments. Jingjing Li, Hongliang Cheng, Yingying Fu, and Jiaming Li evaluated receptor-binding specificity. Zhongyi Wang and Yifei Jin analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
ACKNOWLEDGMENTS
This work was sponsored by the Beijing Nova Program (Z211100002121064) and Fujian Province Health Science and Technology Project (2020CXB050).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
This study was conducted in strict accordance with the recommendations in the “Guidelines for the Care and Use of Laboratory Animals” by the Ministry of Science and Technology of China. The protocol was approved by the Animal Experiment Ethics Committee of Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences (approval number: SCXK 20210166). All virus experiments were performed in biosafety laboratories in the Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Open Research
DATA AVAILABILITY STATEMENT
The data supporting the findings of the current study are available from the corresponding author upon reasonable request.




